Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
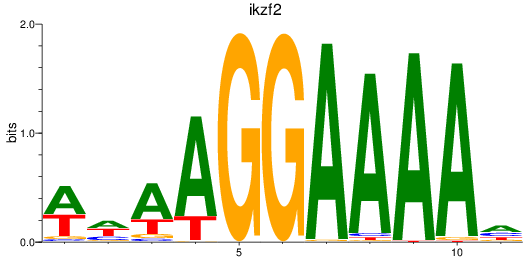
Results for ikzf2
Z-value: 0.69
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr11_v1_chr9_-_40011673_40011673 | -0.38 | 3.1e-01 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23398369 | 1.47 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr16_+_23397785 | 1.43 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr3_+_30921246 | 1.41 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr3_+_55131289 | 1.07 |
ENSDART00000111585
|
AL935210.1
|
|
| chr7_-_35432901 | 1.00 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr11_+_21910343 | 0.95 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr3_-_61203203 | 0.91 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr3_-_20091964 | 0.86 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr3_-_46817838 | 0.82 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_-_30844304 | 0.81 |
ENSDART00000146367
|
crygmxl2
|
crystallin, gamma MX, like 2 |
| chr3_-_46818001 | 0.81 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_48428713 | 0.78 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr21_-_26495700 | 0.74 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr3_-_46817499 | 0.71 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_-_4070058 | 0.70 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr16_+_19732543 | 0.66 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr21_+_25187210 | 0.66 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr1_-_56223913 | 0.64 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr13_-_16257848 | 0.64 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr6_+_39360377 | 0.64 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr3_-_61181018 | 0.64 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr6_+_15250672 | 0.61 |
ENSDART00000155951
|
si:ch73-23l24.1
|
si:ch73-23l24.1 |
| chr1_+_7679328 | 0.60 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr13_+_22264914 | 0.60 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr18_+_19648275 | 0.59 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr9_+_53276356 | 0.59 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr2_+_55984788 | 0.58 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr19_+_48176745 | 0.57 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr6_+_29791164 | 0.56 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr16_+_26774182 | 0.55 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr12_+_28367557 | 0.54 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr12_+_17106117 | 0.53 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr2_-_37210397 | 0.52 |
ENSDART00000084938
|
apoda.1
|
apolipoprotein Da, duplicate 1 |
| chr5_-_68916623 | 0.51 |
ENSDART00000141917
ENSDART00000109053 |
ank1a
|
ankyrin 1, erythrocytic a |
| chr11_+_21910752 | 0.50 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr21_+_11468934 | 0.49 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr13_+_24662238 | 0.49 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr3_+_14611299 | 0.48 |
ENSDART00000140577
|
tspan35
|
tetraspanin 35 |
| chr15_+_29088426 | 0.48 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr16_+_33655890 | 0.47 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr12_+_42436920 | 0.46 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr19_+_30662529 | 0.46 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr11_-_32723851 | 0.46 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr2_+_29976419 | 0.46 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr1_-_40911332 | 0.46 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr15_-_19250543 | 0.44 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr7_+_71664624 | 0.44 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr17_+_52822422 | 0.44 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_-_9980402 | 0.44 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr9_-_22831836 | 0.44 |
ENSDART00000142585
|
neb
|
nebulin |
| chr5_+_4332220 | 0.43 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr15_-_21132480 | 0.43 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr7_+_39386982 | 0.42 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr9_-_1949915 | 0.42 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr19_+_25649626 | 0.42 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr2_+_22042745 | 0.41 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr5_+_36932718 | 0.41 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr18_+_48423973 | 0.40 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_+_13693722 | 0.40 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr25_-_18470695 | 0.40 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr10_-_7785930 | 0.39 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr14_-_3381303 | 0.39 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr4_+_5249494 | 0.39 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr24_+_9744012 | 0.39 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr24_-_23320223 | 0.39 |
ENSDART00000135846
|
zfhx4
|
zinc finger homeobox 4 |
| chr20_+_26702377 | 0.38 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr1_+_1689775 | 0.38 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr4_+_7508316 | 0.38 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr15_+_46356879 | 0.38 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr9_-_8454060 | 0.37 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr7_-_40993456 | 0.37 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr5_-_13835461 | 0.37 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr25_+_21832938 | 0.37 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr12_-_48477031 | 0.37 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr18_+_8346920 | 0.37 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr4_+_9669717 | 0.36 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr10_+_21758811 | 0.36 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr23_-_20764227 | 0.36 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr22_+_20427170 | 0.35 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr2_-_24022085 | 0.35 |
ENSDART00000088643
ENSDART00000189822 |
col15a1b
|
collagen, type XV, alpha 1b |
| chr19_-_31522625 | 0.35 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr14_-_21219659 | 0.35 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr2_-_48196092 | 0.34 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr2_-_22535 | 0.34 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr15_-_21165237 | 0.34 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr22_+_5106751 | 0.34 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr7_+_25920792 | 0.33 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr24_-_26304386 | 0.33 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr20_-_5291012 | 0.33 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr11_+_34235372 | 0.33 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr8_+_18624658 | 0.33 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr4_+_19535946 | 0.33 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr5_-_48260145 | 0.32 |
ENSDART00000044083
ENSDART00000163250 ENSDART00000135911 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr22_-_15602593 | 0.32 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr18_+_26422124 | 0.32 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr13_-_31452516 | 0.32 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr14_-_43000836 | 0.32 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr7_+_65673885 | 0.32 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr12_+_30168342 | 0.32 |
ENSDART00000142756
|
ablim1b
|
actin binding LIM protein 1b |
| chr23_+_36087219 | 0.32 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr16_+_29492937 | 0.32 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr15_+_1372343 | 0.31 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr12_-_431249 | 0.31 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr23_-_21446985 | 0.31 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr7_+_57866292 | 0.31 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr25_+_5039050 | 0.31 |
ENSDART00000154700
|
parvb
|
parvin, beta |
| chr5_-_68916455 | 0.31 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr25_+_3677650 | 0.31 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr4_+_21129752 | 0.30 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr6_+_28124393 | 0.30 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr9_+_5862040 | 0.30 |
ENSDART00000129117
|
pdzk1
|
PDZ domain containing 1 |
| chr20_-_54208344 | 0.30 |
ENSDART00000169386
|
faua
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a |
| chr8_+_10304981 | 0.30 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr6_+_35362225 | 0.29 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr20_+_46040666 | 0.29 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr4_+_15968483 | 0.29 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr1_+_44911405 | 0.29 |
ENSDART00000182465
|
wu:fc21g02
|
wu:fc21g02 |
| chr2_-_30693742 | 0.29 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr2_-_30055432 | 0.29 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr9_+_45839260 | 0.28 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr1_+_9290103 | 0.28 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr9_-_12424791 | 0.28 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr18_-_16123222 | 0.28 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr13_+_15004398 | 0.28 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr12_+_13164706 | 0.27 |
ENSDART00000112565
|
paqr4b
|
progestin and adipoQ receptor family member IVb |
| chr12_+_24344611 | 0.27 |
ENSDART00000093094
|
nrxn1a
|
neurexin 1a |
| chr8_+_1769475 | 0.27 |
ENSDART00000079073
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr22_-_15602760 | 0.27 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr18_+_17663898 | 0.27 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr10_+_21737745 | 0.27 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr24_-_11821505 | 0.26 |
ENSDART00000058992
|
gli3
|
GLI family zinc finger 3 |
| chr10_-_44341288 | 0.26 |
ENSDART00000166131
|
zswim6
|
zinc finger, SWIM-type containing 6 |
| chr23_+_36101185 | 0.26 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr19_-_26869103 | 0.26 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr23_-_24146591 | 0.26 |
ENSDART00000133269
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr8_-_47785951 | 0.26 |
ENSDART00000143821
|
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr18_+_29402623 | 0.26 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr3_-_28428198 | 0.26 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr4_+_4232562 | 0.26 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr8_-_31062811 | 0.26 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr23_+_36106790 | 0.26 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr12_-_15205087 | 0.26 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr18_-_17485419 | 0.26 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr11_-_25418856 | 0.25 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr1_-_26292897 | 0.25 |
ENSDART00000112899
ENSDART00000185410 |
cxxc4
|
CXXC finger 4 |
| chr22_+_6677429 | 0.25 |
ENSDART00000147482
|
si:ch211-209l18.2
|
si:ch211-209l18.2 |
| chr20_+_23173710 | 0.25 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr2_-_31634978 | 0.25 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr3_-_30118856 | 0.25 |
ENSDART00000109953
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr21_+_29077509 | 0.25 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr14_-_21064199 | 0.25 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr23_+_25428513 | 0.25 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr10_+_28587446 | 0.25 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr13_+_1575276 | 0.24 |
ENSDART00000165987
|
DST
|
dystonin |
| chr3_+_37707432 | 0.24 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr10_-_36808348 | 0.24 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr18_+_7591381 | 0.24 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr1_-_669717 | 0.24 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr6_-_18959036 | 0.24 |
ENSDART00000185372
|
sept9b
|
septin 9b |
| chr8_+_15239549 | 0.24 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr21_+_26657404 | 0.24 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr8_-_21372446 | 0.23 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr14_-_14566417 | 0.23 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr21_+_5800306 | 0.23 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr4_-_22207873 | 0.23 |
ENSDART00000142140
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_+_24277556 | 0.23 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr25_-_30357027 | 0.23 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr22_-_20011476 | 0.23 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr3_+_23691847 | 0.23 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr21_+_17880511 | 0.23 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr13_-_31647323 | 0.23 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr4_+_3980247 | 0.23 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr21_+_11468642 | 0.23 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr5_-_38506981 | 0.22 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr12_+_18458502 | 0.22 |
ENSDART00000108745
|
rnf151
|
ring finger protein 151 |
| chr2_-_7696503 | 0.22 |
ENSDART00000169709
|
CABZ01055306.1
|
|
| chr5_-_34609337 | 0.22 |
ENSDART00000145792
|
hexb
|
hexosaminidase B (beta polypeptide) |
| chr17_+_33340675 | 0.22 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr7_+_20524064 | 0.22 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr12_+_24344963 | 0.22 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr16_+_29492749 | 0.22 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr8_-_14049404 | 0.21 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr21_-_33851877 | 0.21 |
ENSDART00000136729
|
ebf1b
|
early B cell factor 1b |
| chr18_+_783936 | 0.21 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr9_+_21407987 | 0.21 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr17_-_14836320 | 0.21 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr7_+_69470442 | 0.21 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr18_+_39074139 | 0.21 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr15_-_46779934 | 0.21 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr3_-_37759373 | 0.21 |
ENSDART00000150962
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr8_+_20880848 | 0.21 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr23_+_44883805 | 0.21 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr16_-_31976269 | 0.21 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr14_+_33413980 | 0.21 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr10_+_9159279 | 0.20 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr4_+_61995745 | 0.20 |
ENSDART00000171539
|
CT990567.1
|
|
| chr21_-_20711739 | 0.20 |
ENSDART00000190918
|
BX530077.1
|
|
| chr1_+_15137901 | 0.20 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr16_-_31756859 | 0.20 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr19_-_27776649 | 0.20 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.4 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.3 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.3 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 1.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.2 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.1 | 0.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.2 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.4 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 0.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.4 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.5 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 3.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.2 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |