Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
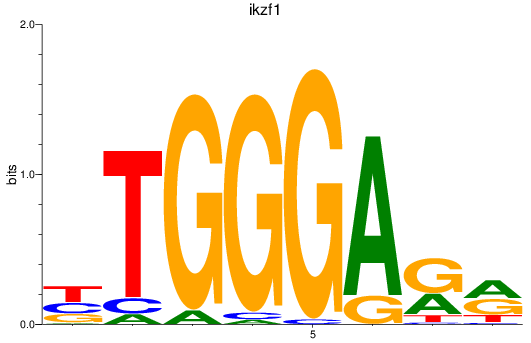
Results for ikzf1
Z-value: 2.04
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr11_v1_chr13_-_15982707_15982707 | 0.73 | 2.7e-02 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32817274 | 3.50 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_-_71722257 | 3.28 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr25_+_29161609 | 3.11 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr7_-_8416750 | 2.84 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr11_-_21030070 | 2.77 |
ENSDART00000186322
|
fmoda
|
fibromodulin a |
| chr5_+_32345187 | 2.48 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr5_+_32222303 | 2.47 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr12_+_25085751 | 2.43 |
ENSDART00000170466
|
gch2
|
GTP cyclohydrolase 2 |
| chr21_+_7582036 | 2.28 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr12_-_16636627 | 2.22 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr20_+_20637866 | 2.21 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr16_+_20915319 | 2.16 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr5_-_28606916 | 2.00 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr7_-_24472991 | 1.99 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr7_-_2039060 | 1.95 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr13_-_39159810 | 1.92 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr20_+_20638034 | 1.92 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr12_-_16720196 | 1.90 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr25_+_18563476 | 1.86 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr21_-_25741096 | 1.86 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr17_+_25414033 | 1.81 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr1_+_135903 | 1.73 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr3_+_30921246 | 1.72 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr6_-_39764995 | 1.71 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_-_35432901 | 1.71 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr13_-_39160018 | 1.69 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr17_+_23300827 | 1.69 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr7_-_8438657 | 1.68 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr2_+_42724404 | 1.68 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr12_-_16619449 | 1.67 |
ENSDART00000182074
|
ctslb
|
cathepsin Lb |
| chr23_-_26077038 | 1.64 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr15_-_23376541 | 1.59 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr12_-_16595177 | 1.58 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr17_+_2549503 | 1.57 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr13_-_36391496 | 1.57 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr25_-_31396479 | 1.54 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr5_-_41831646 | 1.50 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr18_-_14860435 | 1.48 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr14_-_17068511 | 1.47 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr7_-_58098814 | 1.45 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr12_+_42574148 | 1.44 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr2_-_31936966 | 1.43 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr21_+_19834072 | 1.42 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr18_+_30847237 | 1.42 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr10_+_21786656 | 1.39 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr7_-_8490886 | 1.37 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr6_+_6491013 | 1.36 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr5_-_31712399 | 1.35 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr14_-_51855047 | 1.34 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr10_-_8358396 | 1.34 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr16_-_31717851 | 1.29 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr1_+_51496862 | 1.28 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr17_+_10242166 | 1.28 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr1_-_44434707 | 1.28 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr16_+_1353894 | 1.25 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr7_-_32833153 | 1.25 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr1_+_47446032 | 1.25 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr16_-_46660680 | 1.24 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr21_-_10773344 | 1.23 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr15_+_36115955 | 1.23 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr15_-_24869826 | 1.22 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr25_-_15045338 | 1.22 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr5_-_8164439 | 1.22 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr7_+_31879649 | 1.21 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr5_-_64883082 | 1.18 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr23_-_11870962 | 1.18 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr5_+_72108241 | 1.15 |
ENSDART00000006606
|
fabp1a
|
fatty acid binding protein 1a, liver |
| chr3_+_25154078 | 1.14 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr1_+_36436936 | 1.14 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr16_-_31718013 | 1.13 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr14_-_5678457 | 1.11 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_-_43787616 | 1.09 |
ENSDART00000179758
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_+_37391141 | 1.09 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr17_+_996509 | 1.08 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr17_+_52822422 | 1.08 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr19_+_12583577 | 1.08 |
ENSDART00000151508
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr14_-_39031108 | 1.08 |
ENSDART00000026194
|
glra4a
|
glycine receptor, alpha 4a |
| chr2_-_21335131 | 1.08 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr7_+_39386982 | 1.03 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr5_-_26181863 | 1.03 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr15_-_1822166 | 1.02 |
ENSDART00000156883
|
mmp28
|
matrix metallopeptidase 28 |
| chr19_-_47452874 | 1.02 |
ENSDART00000025931
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr20_+_54738210 | 1.01 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr23_-_30787932 | 1.00 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr11_+_41981959 | 1.00 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr2_-_34555945 | 1.00 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr23_+_36083529 | 1.00 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr25_-_23526058 | 0.98 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr23_+_6795531 | 0.98 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr9_-_21067971 | 0.97 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr18_+_20494413 | 0.96 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr16_+_5156420 | 0.96 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr9_-_1939232 | 0.96 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr10_+_22724059 | 0.95 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr14_-_17068712 | 0.93 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr1_+_36437585 | 0.93 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr2_-_1518360 | 0.92 |
ENSDART00000182788
|
c8b
|
complement component 8, beta polypeptide |
| chr15_-_2632891 | 0.92 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr10_-_25860102 | 0.90 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr9_+_21402863 | 0.89 |
ENSDART00000125357
|
cx30.3
|
connexin 30.3 |
| chr19_+_22216778 | 0.89 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr13_+_27951688 | 0.89 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr12_-_3940768 | 0.89 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr16_+_50100420 | 0.88 |
ENSDART00000128167
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr9_-_5046315 | 0.88 |
ENSDART00000179087
ENSDART00000109954 |
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr15_-_2640966 | 0.86 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr18_-_41375120 | 0.85 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr7_+_8456999 | 0.85 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr3_+_40170216 | 0.84 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr23_+_23658474 | 0.84 |
ENSDART00000162838
|
agrn
|
agrin |
| chr3_+_41922114 | 0.84 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_-_41841675 | 0.84 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr1_-_46981134 | 0.83 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_-_4925641 | 0.82 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr16_-_54455573 | 0.82 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr3_-_58644920 | 0.80 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr5_-_14344647 | 0.79 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr5_+_32791245 | 0.79 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr18_+_45990394 | 0.79 |
ENSDART00000024068
|
mmp23bb
|
matrix metallopeptidase 23bb |
| chr19_-_9829965 | 0.78 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr20_+_1398564 | 0.78 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr6_+_39370587 | 0.78 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr12_+_36971952 | 0.78 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr16_+_29492937 | 0.77 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr24_-_38816725 | 0.77 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr4_+_25651720 | 0.77 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr18_-_7400075 | 0.76 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr16_+_37470717 | 0.76 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr4_-_12914163 | 0.75 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr5_-_41838354 | 0.75 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr8_-_18535822 | 0.75 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr18_-_7399767 | 0.75 |
ENSDART00000181689
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr5_-_26466169 | 0.74 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr23_+_2361184 | 0.73 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr18_+_45796096 | 0.73 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr12_-_30548244 | 0.71 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr18_-_15911394 | 0.70 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr19_+_12801940 | 0.70 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr14_-_2196267 | 0.69 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr8_-_34762163 | 0.69 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr4_-_68913650 | 0.68 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr13_-_46991577 | 0.68 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr2_-_689047 | 0.67 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr21_+_39100289 | 0.65 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr7_-_8470860 | 0.64 |
ENSDART00000172793
|
loc564481
|
hypothetical protein LOC564481 |
| chr2_-_10338759 | 0.64 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr17_+_32374876 | 0.64 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr21_-_131236 | 0.64 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr17_-_14876758 | 0.64 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr15_+_42573909 | 0.64 |
ENSDART00000181801
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr23_+_3616224 | 0.64 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr24_-_2900511 | 0.63 |
ENSDART00000185674
|
fam69c
|
family with sequence similarity 69, member C |
| chr23_+_21663631 | 0.63 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr9_+_33145522 | 0.63 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr12_+_31673588 | 0.63 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr15_-_2652640 | 0.63 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr7_+_33314925 | 0.63 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr15_-_28247583 | 0.63 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr5_-_57641257 | 0.62 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr11_+_10541258 | 0.62 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr16_+_24842662 | 0.61 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr8_+_1009831 | 0.61 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr2_+_6885852 | 0.60 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr20_+_2039518 | 0.60 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr8_+_20488322 | 0.60 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr23_-_3674443 | 0.58 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr13_-_37102522 | 0.58 |
ENSDART00000147884
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr21_-_40835069 | 0.58 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr21_-_12119711 | 0.57 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr2_+_25278107 | 0.57 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr20_+_43942278 | 0.57 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr15_-_20233105 | 0.57 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr1_-_35916247 | 0.56 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr11_-_277599 | 0.56 |
ENSDART00000187109
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr21_+_19648814 | 0.56 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr16_-_29437373 | 0.56 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr6_+_18142623 | 0.56 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr12_+_17100021 | 0.56 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr15_-_12229874 | 0.55 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr20_+_320415 | 0.55 |
ENSDART00000152344
|
si:dkey-119m7.8
|
si:dkey-119m7.8 |
| chr15_-_18176694 | 0.55 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr8_+_52642869 | 0.55 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr20_+_2240278 | 0.54 |
ENSDART00000041250
|
tmem200a
|
transmembrane protein 200A |
| chr8_+_6967108 | 0.54 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr13_+_38302665 | 0.54 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr15_+_1796313 | 0.54 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr25_+_10830269 | 0.53 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr23_-_24582606 | 0.52 |
ENSDART00000129910
|
tmem240a
|
transmembrane protein 240a |
| chr2_-_32501501 | 0.52 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr9_-_11549379 | 0.52 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr6_+_48618512 | 0.51 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr15_-_8517376 | 0.51 |
ENSDART00000186289
|
npas1
|
neuronal PAS domain protein 1 |
| chr17_-_48705993 | 0.51 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr21_+_15824182 | 0.51 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr23_-_18287618 | 0.51 |
ENSDART00000112735
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr1_-_206208 | 0.50 |
ENSDART00000060968
|
adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr7_+_36898622 | 0.50 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr7_+_57866292 | 0.50 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr9_-_21067673 | 0.50 |
ENSDART00000180257
|
tbx15
|
T-box 15 |
| chr5_-_41841892 | 0.49 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 2.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.6 | 2.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.6 | 2.4 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.5 | 3.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.4 | 2.3 | GO:0035176 | mammillary body development(GO:0021767) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 1.9 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.4 | 1.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 1.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 1.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 0.8 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 1.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 2.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.8 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.2 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.9 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.2 | 0.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 0.7 | GO:0048241 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.2 | 0.5 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.7 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.2 | 0.8 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 1.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 1.7 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 1.2 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 3.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.9 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 2.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.4 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.7 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.4 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 3.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.2 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.8 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.6 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.3 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:1901021 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of calcium ion transmembrane transport(GO:1904427) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0003416 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.4 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.2 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.5 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 2.5 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 1.6 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.6 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 4.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 5.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 26.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.8 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.4 | 1.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 3.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.6 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 2.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.5 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.2 | 0.5 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 1.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 1.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 1.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.2 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 4.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.5 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 1.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 3.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |