Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
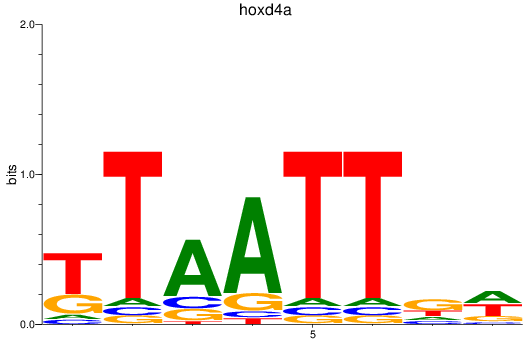
Results for hoxd4a
Z-value: 0.71
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr11_v1_chr9_-_1951144_1951144 | -0.65 | 6.1e-02 | Click! |
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2578026 | 1.04 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_9985019 | 0.88 |
ENSDART00000193536
ENSDART00000189595 |
zgc:171977
|
zgc:171977 |
| chr7_-_20582842 | 0.80 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr24_-_9979342 | 0.75 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr1_-_18811517 | 0.74 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr5_-_68795063 | 0.73 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr18_-_33979693 | 0.66 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr10_-_26202766 | 0.64 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr16_+_29509133 | 0.57 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_-_37973819 | 0.55 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr17_-_4252221 | 0.50 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr6_+_4387150 | 0.47 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr11_+_18183220 | 0.44 |
ENSDART00000113468
|
LO018315.10
|
|
| chr1_-_45213565 | 0.44 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr7_+_44802353 | 0.43 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr14_-_33481428 | 0.42 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr13_-_37631092 | 0.40 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr14_-_30905288 | 0.39 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr21_+_4256291 | 0.38 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr7_-_26532089 | 0.38 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr9_+_29548195 | 0.37 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr21_-_42831033 | 0.37 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr24_-_11506054 | 0.36 |
ENSDART00000140217
ENSDART00000106310 |
prpf4bb
|
pre-mRNA processing factor 4Bb |
| chr24_-_30862168 | 0.36 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr19_-_3167729 | 0.35 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr5_-_1869982 | 0.35 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr8_+_23783066 | 0.35 |
ENSDART00000062968
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr15_+_42397125 | 0.34 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr18_-_5392667 | 0.34 |
ENSDART00000169896
|
CABZ01084603.1
|
|
| chr11_+_42641404 | 0.33 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr13_+_2861265 | 0.33 |
ENSDART00000170602
ENSDART00000171687 |
XPO5
|
si:ch211-233m11.2 |
| chr3_-_32873641 | 0.33 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr6_+_49053319 | 0.32 |
ENSDART00000124524
|
sycp1
|
synaptonemal complex protein 1 |
| chr23_-_16980213 | 0.32 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr14_+_26247319 | 0.31 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr2_+_33326522 | 0.31 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr22_-_26289549 | 0.31 |
ENSDART00000043774
|
sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr2_-_23677422 | 0.30 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr4_+_13586455 | 0.30 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr23_+_19590006 | 0.29 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr8_-_29930821 | 0.29 |
ENSDART00000125173
|
ercc6l2
|
excision repair cross-complementation group 6-like 2 |
| chr19_+_45962016 | 0.28 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr20_-_51186524 | 0.28 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr4_+_65127317 | 0.28 |
ENSDART00000166475
|
znf1126
|
zinc finger protein 1126 |
| chr22_+_25774750 | 0.28 |
ENSDART00000174421
|
AL929192.1
|
|
| chr11_-_35171768 | 0.28 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr23_-_31913069 | 0.27 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr10_+_39893439 | 0.27 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr6_+_36839509 | 0.26 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr9_+_38457806 | 0.26 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr10_-_44355534 | 0.26 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr25_+_30074947 | 0.26 |
ENSDART00000154849
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr5_+_67268110 | 0.25 |
ENSDART00000148116
|
si:ch211-110p13.9
|
si:ch211-110p13.9 |
| chr4_-_65525733 | 0.25 |
ENSDART00000159401
|
BX293993.1
|
|
| chr7_+_6941583 | 0.25 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr6_+_3693441 | 0.25 |
ENSDART00000065256
|
ppig
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr2_+_16696052 | 0.24 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr24_-_2450597 | 0.24 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr15_+_42285643 | 0.24 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr18_-_17147803 | 0.24 |
ENSDART00000187986
ENSDART00000169080 |
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr5_+_26847190 | 0.23 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr23_-_19051710 | 0.23 |
ENSDART00000111852
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr7_-_73851280 | 0.23 |
ENSDART00000190053
|
FP236812.3
|
|
| chr2_-_39558643 | 0.23 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr4_+_17671164 | 0.22 |
ENSDART00000040445
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr1_+_50921266 | 0.22 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr13_+_33368140 | 0.22 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr11_+_1608348 | 0.22 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr18_+_13077800 | 0.22 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr24_+_26134209 | 0.22 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr7_+_5910467 | 0.22 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr2_+_36121373 | 0.22 |
ENSDART00000187002
|
CT867973.2
|
|
| chr16_+_13818743 | 0.21 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr17_+_21477892 | 0.21 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr1_+_54655160 | 0.21 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr12_+_25223843 | 0.21 |
ENSDART00000077180
ENSDART00000127454 ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr2_+_20793982 | 0.21 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr17_+_30843881 | 0.21 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr17_-_13070602 | 0.21 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr7_+_56676838 | 0.21 |
ENSDART00000112242
|
zgc:194679
|
zgc:194679 |
| chr7_-_6446172 | 0.20 |
ENSDART00000173212
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr11_+_24034345 | 0.20 |
ENSDART00000128309
|
zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr22_+_7439186 | 0.20 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr11_-_22303678 | 0.20 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr23_-_19051869 | 0.20 |
ENSDART00000140866
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr17_-_14559576 | 0.20 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr16_+_26017360 | 0.20 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr11_-_44979281 | 0.20 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr13_-_49585385 | 0.20 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr23_-_29553430 | 0.19 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr18_+_3572314 | 0.19 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr4_-_68755773 | 0.19 |
ENSDART00000163119
|
znf985
|
zinc finger protein 985 |
| chr7_-_2129076 | 0.19 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr2_+_56657804 | 0.19 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr11_+_11303458 | 0.19 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr23_-_31913231 | 0.19 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr18_-_48296793 | 0.19 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr21_+_21195487 | 0.19 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr2_-_38206034 | 0.18 |
ENSDART00000144518
ENSDART00000137395 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr2_+_23677179 | 0.18 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr12_-_2869565 | 0.18 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr16_+_9726038 | 0.18 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr16_+_25107344 | 0.18 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr18_+_44849809 | 0.17 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr25_-_2723682 | 0.17 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr23_+_2666944 | 0.17 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr19_-_43757568 | 0.17 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr4_-_55592720 | 0.17 |
ENSDART00000164747
|
znf1029
|
zinc finger protein 1029 |
| chr7_-_41512999 | 0.17 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr2_-_1364678 | 0.17 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr7_+_22657566 | 0.17 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr6_-_40922971 | 0.17 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr1_-_999556 | 0.16 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr10_-_24759616 | 0.16 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr22_+_16293071 | 0.16 |
ENSDART00000170960
|
dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr5_-_61970674 | 0.16 |
ENSDART00000143161
|
znf541
|
zinc finger protein 541 |
| chr15_-_14884332 | 0.16 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr4_+_9028819 | 0.16 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr17_+_13088800 | 0.16 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr5_-_18924834 | 0.16 |
ENSDART00000041371
|
poli
|
polymerase (DNA directed) iota |
| chr1_-_55044256 | 0.15 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr8_+_22377770 | 0.15 |
ENSDART00000132841
|
si:dkey-23c22.5
|
si:dkey-23c22.5 |
| chr11_+_44804685 | 0.15 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr14_-_31618243 | 0.15 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr21_+_13383413 | 0.15 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr22_-_2922053 | 0.15 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr1_+_44003654 | 0.15 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr5_-_31856681 | 0.14 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr9_-_27720612 | 0.14 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr16_-_10837245 | 0.14 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr6_+_39098397 | 0.14 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr2_+_20802876 | 0.14 |
ENSDART00000142321
|
prg4a
|
proteoglycan 4a |
| chr16_-_47427016 | 0.14 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr14_+_22022441 | 0.14 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr25_-_27729046 | 0.14 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr5_+_18925367 | 0.14 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr14_-_31489410 | 0.14 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr12_+_18899396 | 0.14 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr4_-_70883663 | 0.14 |
ENSDART00000170959
|
si:ch211-108c17.2
|
si:ch211-108c17.2 |
| chr6_+_36381709 | 0.14 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr9_+_34380299 | 0.13 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr9_+_28150275 | 0.13 |
ENSDART00000192129
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr8_+_8238727 | 0.13 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr22_-_24757785 | 0.13 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr7_-_59311165 | 0.13 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr5_-_37871526 | 0.12 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr15_-_34658057 | 0.12 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr5_+_27897504 | 0.12 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr23_+_2760573 | 0.12 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr7_+_12927944 | 0.12 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr2_+_33726862 | 0.12 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr11_-_31146808 | 0.12 |
ENSDART00000172781
|
si:cabz01069582.1
|
si:cabz01069582.1 |
| chr15_-_28082310 | 0.11 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr10_+_40660772 | 0.11 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr11_+_1602916 | 0.11 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr11_+_15878343 | 0.11 |
ENSDART00000167191
ENSDART00000171862 ENSDART00000163992 ENSDART00000170065 |
pank4
|
pantothenate kinase 4 |
| chr16_+_25074029 | 0.11 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr6_+_41191482 | 0.11 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr8_+_26230060 | 0.11 |
ENSDART00000155529
ENSDART00000140798 |
wdr6
|
WD repeat domain 6 |
| chr13_+_48359573 | 0.11 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr5_+_24520437 | 0.11 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr18_-_33979422 | 0.10 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr23_-_1017605 | 0.10 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr1_+_43686251 | 0.10 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr12_+_30367079 | 0.10 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr2_+_2181709 | 0.10 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr12_+_47698356 | 0.10 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr13_+_9468535 | 0.10 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr20_-_19511700 | 0.10 |
ENSDART00000040191
|
snx17
|
sorting nexin 17 |
| chr25_-_1348370 | 0.10 |
ENSDART00000154865
|
itga11b
|
integrin, alpha 11b |
| chr23_-_24343363 | 0.10 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr17_-_28097760 | 0.10 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr22_-_18241390 | 0.09 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr23_-_16485190 | 0.09 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr25_+_29474583 | 0.09 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr25_-_2723902 | 0.09 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr24_+_39638555 | 0.09 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr9_+_31534601 | 0.09 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr16_-_13004166 | 0.09 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr8_-_50888806 | 0.09 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr13_-_4018888 | 0.09 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr1_-_46875493 | 0.09 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_-_9873652 | 0.09 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr6_+_21992820 | 0.09 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr5_+_36611128 | 0.09 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr18_+_13309862 | 0.08 |
ENSDART00000131555
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr2_+_4146299 | 0.08 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr10_-_40484531 | 0.08 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr4_-_14915268 | 0.08 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr4_-_11098539 | 0.08 |
ENSDART00000135511
|
si:dkey-21h14.12
|
si:dkey-21h14.12 |
| chr22_+_37631034 | 0.08 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr22_+_18166660 | 0.08 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr9_+_48219111 | 0.08 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr2_-_47923803 | 0.08 |
ENSDART00000056306
|
ftr24
|
finTRIM family, member 24 |
| chr20_-_29633507 | 0.07 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr13_+_40034176 | 0.07 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr20_+_36628059 | 0.07 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr3_+_52275691 | 0.07 |
ENSDART00000190109
|
BX511132.2
|
|
| chr3_-_49382896 | 0.07 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr6_+_8626427 | 0.07 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.5 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.0 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.4 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.0 | 0.1 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.5 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.4 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.9 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.4 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |