Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for hoxd3a
Z-value: 0.47
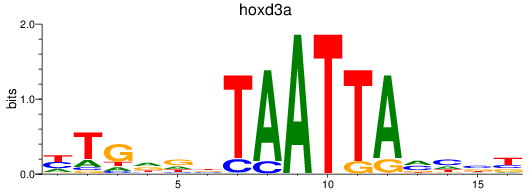
Transcription factors associated with hoxd3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd3a
|
ENSDARG00000059280 | homeobox D3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd3a | dr11_v1_chr9_-_1949915_1949915 | 0.59 | 9.8e-02 | Click! |
Activity profile of hoxd3a motif
Sorted Z-values of hoxd3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 0.91 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_+_52392511 | 0.77 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr10_+_39084354 | 0.76 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr7_-_48667056 | 0.58 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr17_+_23298928 | 0.56 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr9_-_21918963 | 0.52 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr20_-_7072487 | 0.51 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr18_+_3037998 | 0.46 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr25_+_29160102 | 0.43 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr1_-_58036509 | 0.41 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr8_-_31107537 | 0.39 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr3_+_30500968 | 0.38 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr4_+_58576146 | 0.38 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr12_+_24952902 | 0.37 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr1_-_40911332 | 0.36 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr8_+_17078692 | 0.36 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr15_-_35252522 | 0.36 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr20_+_539852 | 0.34 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr15_+_5360407 | 0.32 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr13_-_31435137 | 0.32 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr11_-_40728380 | 0.32 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr16_+_5251768 | 0.31 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr10_+_39091353 | 0.31 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr16_+_31804590 | 0.28 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr10_-_26744131 | 0.28 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr19_-_27462720 | 0.26 |
ENSDART00000135901
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr3_+_27798094 | 0.25 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr14_+_5385855 | 0.23 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr3_-_34084387 | 0.22 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr16_+_14707960 | 0.22 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr4_+_47636303 | 0.22 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr22_-_10117918 | 0.22 |
ENSDART00000177475
ENSDART00000128590 |
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr1_-_50859053 | 0.21 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr12_+_30265802 | 0.21 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr10_+_39091716 | 0.21 |
ENSDART00000193072
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr23_+_579893 | 0.19 |
ENSDART00000189098
|
BX323461.1
|
|
| chr8_+_22405477 | 0.18 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr20_-_14925281 | 0.18 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr20_-_46362606 | 0.16 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr19_-_6988837 | 0.16 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr20_-_14924858 | 0.15 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr8_-_50888806 | 0.13 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr14_+_22591624 | 0.12 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr7_+_24165371 | 0.11 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr21_-_35419486 | 0.11 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr1_+_56755536 | 0.10 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr5_-_38094130 | 0.09 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr10_-_41400049 | 0.08 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr4_-_71210030 | 0.08 |
ENSDART00000186473
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr8_-_30979494 | 0.08 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr22_-_209741 | 0.08 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr6_+_41191482 | 0.07 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr10_+_11355841 | 0.07 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr17_-_37054959 | 0.07 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr10_+_43039947 | 0.07 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr12_+_22580579 | 0.06 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_-_10971230 | 0.06 |
ENSDART00000166196
|
LO018584.1
|
|
| chr19_-_14155781 | 0.05 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr21_-_37790727 | 0.05 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr5_+_66353589 | 0.05 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr15_+_31899312 | 0.05 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr8_+_25034544 | 0.04 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr5_-_3684402 | 0.04 |
ENSDART00000184354
|
FO704648.1
|
|
| chr14_+_30762131 | 0.03 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr4_+_73085993 | 0.03 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr18_+_9362455 | 0.03 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr13_-_44630111 | 0.03 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr20_-_9095105 | 0.02 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_-_40029423 | 0.02 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr18_+_17827149 | 0.02 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr10_-_5847655 | 0.02 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr9_+_22364997 | 0.01 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr19_+_11217279 | 0.01 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr11_+_2391469 | 0.01 |
ENSDART00000182121
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr5_+_28160503 | 0.01 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr19_-_7690975 | 0.00 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.6 | GO:0016102 | diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |