Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for hoxd10a
Z-value: 0.69
Transcription factors associated with hoxd10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd10a
|
ENSDARG00000057859 | homeobox D10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd10a | dr11_v1_chr9_-_1970071_1970071 | -0.93 | 2.6e-04 | Click! |
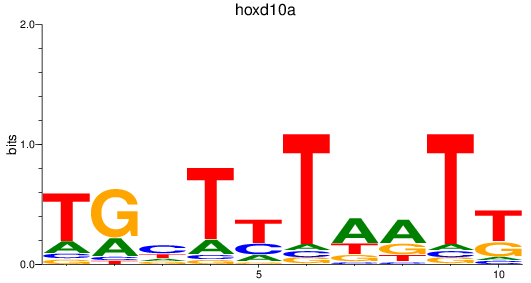
Activity profile of hoxd10a motif
Sorted Z-values of hoxd10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_28322986 | 1.52 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr1_-_18811517 | 1.40 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr2_-_1486023 | 1.08 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr6_-_25165693 | 1.03 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr22_-_38819603 | 0.97 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr21_+_5192016 | 0.88 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr11_-_39118882 | 0.82 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr24_+_17069420 | 0.82 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr19_+_13994563 | 0.76 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr3_-_40768548 | 0.75 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr3_-_26191960 | 0.73 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr24_+_15655233 | 0.73 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr9_+_29643036 | 0.72 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr25_-_12412704 | 0.71 |
ENSDART00000168275
|
det1
|
DET1, COP1 ubiquitin ligase partner |
| chr1_-_55068941 | 0.71 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr19_-_3056235 | 0.70 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr5_-_23805387 | 0.69 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr12_+_30788912 | 0.68 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr25_+_18436301 | 0.67 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr24_+_15655069 | 0.66 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr18_-_20458412 | 0.65 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr18_-_39200557 | 0.64 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr25_+_34915762 | 0.63 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr10_+_41986685 | 0.63 |
ENSDART00000086165
ENSDART00000171983 |
setd1ba
|
SET domain containing 1B, a |
| chr22_-_15010688 | 0.62 |
ENSDART00000139892
|
elfn2a
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2a |
| chr21_+_34167178 | 0.60 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr21_+_3796196 | 0.59 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr13_-_31017960 | 0.57 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr2_+_21356242 | 0.57 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr21_+_3796620 | 0.57 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr9_-_19489264 | 0.54 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr9_-_3519717 | 0.53 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr19_-_35450661 | 0.52 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr11_+_14104417 | 0.49 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr11_-_35970966 | 0.49 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr19_+_4051535 | 0.48 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr25_-_3217115 | 0.48 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr19_+_43828104 | 0.48 |
ENSDART00000166031
|
eloa
|
elongin A |
| chr9_+_3519191 | 0.47 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr11_-_27953135 | 0.47 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr9_+_45605410 | 0.46 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr21_+_21279159 | 0.45 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr5_+_6672870 | 0.44 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr25_-_2723682 | 0.43 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr3_-_43954343 | 0.43 |
ENSDART00000157580
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr15_-_10455438 | 0.42 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr10_+_44692272 | 0.42 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr14_+_23184517 | 0.41 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr16_+_41060161 | 0.38 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr1_+_54199406 | 0.38 |
ENSDART00000176578
|
tsc2
|
TSC complex subunit 2 |
| chr14_+_44804326 | 0.36 |
ENSDART00000079866
ENSDART00000172974 |
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr3_-_11828206 | 0.36 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr6_-_39521832 | 0.36 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr13_-_24669258 | 0.36 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr3_+_29600917 | 0.33 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr14_-_32959851 | 0.33 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_+_32341734 | 0.32 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr11_-_23687158 | 0.31 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr6_-_40921412 | 0.31 |
ENSDART00000076097
ENSDART00000188364 |
sfi1
|
SFI1 centrin binding protein |
| chr1_-_19764038 | 0.31 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr13_+_2442841 | 0.31 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr16_-_43233509 | 0.30 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr10_-_2788668 | 0.30 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr5_+_50912729 | 0.28 |
ENSDART00000190837
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr1_+_26105141 | 0.28 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr3_-_26184018 | 0.28 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr6_+_9793791 | 0.28 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr5_+_30986512 | 0.27 |
ENSDART00000005893
|
ube2g1a
|
ubiquitin-conjugating enzyme E2G 1a (UBC7 homolog, yeast) |
| chr14_+_20941038 | 0.27 |
ENSDART00000182539
|
zgc:66433
|
zgc:66433 |
| chr5_+_35458346 | 0.27 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr5_+_22459087 | 0.26 |
ENSDART00000134781
|
BX546499.1
|
|
| chr25_-_2723902 | 0.25 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr13_-_18069421 | 0.25 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr16_-_42186093 | 0.25 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr5_+_10084100 | 0.24 |
ENSDART00000109236
|
si:ch211-207k7.4
|
si:ch211-207k7.4 |
| chr19_-_10324182 | 0.24 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr6_-_14040136 | 0.23 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr18_+_23218980 | 0.23 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr3_-_36246899 | 0.22 |
ENSDART00000141106
ENSDART00000059506 ENSDART00000158701 |
gps1
|
G protein pathway suppressor 1 |
| chr19_-_10324573 | 0.22 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr13_+_24717880 | 0.22 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr25_+_34915576 | 0.20 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr21_-_3770636 | 0.19 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr23_+_12852655 | 0.19 |
ENSDART00000183015
|
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr18_-_42395161 | 0.19 |
ENSDART00000098641
|
cntn5
|
contactin 5 |
| chr8_-_53166975 | 0.17 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr8_-_15292197 | 0.15 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr20_-_25445237 | 0.13 |
ENSDART00000145034
ENSDART00000018049 |
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr1_-_17711636 | 0.12 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr6_+_29386728 | 0.12 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr18_-_2222128 | 0.10 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr23_-_17451264 | 0.10 |
ENSDART00000190748
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr5_+_19094462 | 0.10 |
ENSDART00000190596
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr22_+_22302614 | 0.10 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr20_-_30931139 | 0.09 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr2_-_42628028 | 0.09 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr7_+_17973269 | 0.09 |
ENSDART00000079969
|
kcnk7
|
potassium channel, subfamily K, member 7 |
| chr10_-_3138403 | 0.09 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr9_+_19489304 | 0.09 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr4_-_16824556 | 0.08 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr17_+_31621788 | 0.08 |
ENSDART00000111629
ENSDART00000157148 |
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr9_-_3519253 | 0.07 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr19_+_47299212 | 0.07 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr10_-_26985231 | 0.06 |
ENSDART00000050585
|
znhit2
|
zinc finger, HIT-type containing 2 |
| chr21_-_11654422 | 0.06 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr18_+_15644559 | 0.06 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_+_12316438 | 0.05 |
ENSDART00000156854
|
si:dkey-276j7.2
|
si:dkey-276j7.2 |
| chr22_-_36750589 | 0.05 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
| chr15_+_3674765 | 0.04 |
ENSDART00000081802
|
NEK3
|
NIMA related kinase 3 |
| chr11_-_42752884 | 0.04 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr10_-_36244864 | 0.04 |
ENSDART00000123748
|
or109-1
|
odorant receptor, family D, subfamily 109, member 1 |
| chr20_+_13894123 | 0.03 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr2_-_23349116 | 0.03 |
ENSDART00000099690
|
fam129ab
|
family with sequence similarity 129, member Ab |
| chr17_-_19344999 | 0.02 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr7_+_5007310 | 0.02 |
ENSDART00000146083
|
si:ch211-272h9.3
|
si:ch211-272h9.3 |
| chr5_+_19094153 | 0.02 |
ENSDART00000186525
ENSDART00000064752 |
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr10_-_45379831 | 0.02 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr22_+_21950838 | 0.00 |
ENSDART00000143075
|
gna15.4
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 4 |
| chr23_-_41585748 | 0.00 |
ENSDART00000102879
ENSDART00000188689 |
scube3
|
signal peptide, CUB domain, EGF-like 3 |
| chr18_+_7543347 | 0.00 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.5 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.4 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.7 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 1.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.4 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.8 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0006096 | glycolytic process(GO:0006096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.7 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |