Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
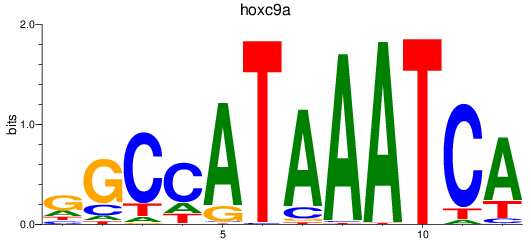
Results for hoxc9a
Z-value: 1.88
Transcription factors associated with hoxc9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc9a
|
ENSDARG00000092809 | homeobox C9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc9a | dr11_v1_chr23_+_36095260_36095260 | 0.84 | 5.1e-03 | Click! |
Activity profile of hoxc9a motif
Sorted Z-values of hoxc9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47633438 | 6.83 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr3_-_61205711 | 5.80 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr6_+_55032439 | 4.30 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr3_+_33300522 | 3.59 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr17_+_23300827 | 3.51 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr5_-_51619262 | 3.21 |
ENSDART00000134606
ENSDART00000081249 |
otpb
|
orthopedia homeobox b |
| chr8_+_16004154 | 2.94 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr13_+_4671698 | 2.83 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr5_-_51619742 | 2.80 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr3_+_40809011 | 2.58 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr19_+_42983613 | 2.46 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr6_+_52804267 | 2.37 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr19_+_19775757 | 2.37 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr5_+_36932718 | 2.29 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr8_+_6954984 | 2.27 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr17_+_32343121 | 2.22 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr13_-_43599898 | 2.18 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr3_+_23742868 | 2.18 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr13_+_15004398 | 2.15 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr7_+_31879649 | 2.12 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr21_-_27338639 | 2.08 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr3_+_23743139 | 2.02 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr2_+_2223837 | 2.00 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr11_-_6188413 | 1.87 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr7_+_31879986 | 1.86 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr8_+_22931427 | 1.86 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr16_-_35952789 | 1.77 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr6_-_46875310 | 1.70 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr19_+_19756425 | 1.68 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr1_+_16144615 | 1.64 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr12_+_16440708 | 1.59 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr19_+_4916233 | 1.59 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr25_+_3058924 | 1.57 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr25_+_14109626 | 1.56 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr17_+_51262556 | 1.54 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr4_+_10017049 | 1.53 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr11_+_16040680 | 1.47 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr23_+_43255328 | 1.41 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr1_-_39943596 | 1.40 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr10_+_20128267 | 1.36 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr10_+_10788811 | 1.30 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr15_-_14375452 | 1.30 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr3_-_12930217 | 1.30 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr7_-_26408472 | 1.29 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr11_+_16040517 | 1.28 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr14_-_17072736 | 1.28 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr17_+_18117029 | 1.27 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr1_-_681116 | 1.25 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr23_+_20669149 | 1.24 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr25_-_13842618 | 1.20 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr4_+_16715267 | 1.18 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr14_-_36799280 | 1.17 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr11_-_3334248 | 1.17 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr14_+_36231126 | 1.13 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr4_+_10754802 | 1.05 |
ENSDART00000181470
|
stab2
|
stabilin 2 |
| chr15_-_5467477 | 0.96 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr25_-_5963535 | 0.95 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr21_-_131236 | 0.94 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr5_-_71838520 | 0.91 |
ENSDART00000174396
|
CU927890.1
|
|
| chr8_-_23701880 | 0.89 |
ENSDART00000139897
|
si:ch73-237c6.1
|
si:ch73-237c6.1 |
| chr10_-_19801821 | 0.84 |
ENSDART00000148013
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr21_+_11969603 | 0.82 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr25_-_1720736 | 0.81 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr10_-_11385155 | 0.79 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr3_+_34821327 | 0.78 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr10_+_26571174 | 0.78 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr10_+_439692 | 0.78 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr2_-_18830722 | 0.77 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr22_-_21046654 | 0.75 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr1_-_39909985 | 0.71 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr7_-_41964877 | 0.66 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr17_-_20849879 | 0.66 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr17_-_19626357 | 0.64 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr10_+_31953502 | 0.64 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr5_-_16351306 | 0.62 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr22_-_21046843 | 0.61 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr5_+_9408901 | 0.50 |
ENSDART00000193364
|
FP236810.1
|
|
| chr21_+_42930558 | 0.47 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr12_+_27213733 | 0.42 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr15_+_8043751 | 0.39 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr12_-_6172154 | 0.39 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr14_+_35024521 | 0.38 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr14_+_11762991 | 0.36 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr13_+_31583034 | 0.35 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr14_+_21783400 | 0.35 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr12_-_20362041 | 0.35 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr18_-_2433011 | 0.35 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr21_-_1742159 | 0.32 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr9_+_16449398 | 0.32 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr20_+_27464721 | 0.32 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr25_+_19008497 | 0.31 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr11_+_19159083 | 0.30 |
ENSDART00000138964
|
adamts9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr17_+_36627099 | 0.29 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr1_-_23157583 | 0.28 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr4_+_11439511 | 0.26 |
ENSDART00000150485
|
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr15_+_8767650 | 0.25 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr14_-_30141162 | 0.24 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr11_+_25044082 | 0.24 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr2_+_42715995 | 0.23 |
ENSDART00000143419
ENSDART00000183914 ENSDART00000184371 ENSDART00000185485 ENSDART00000037332 |
ftr12
|
finTRIM family, member 12 |
| chr3_-_37699992 | 0.23 |
ENSDART00000151193
|
gpatch8
|
G patch domain containing 8 |
| chr17_-_30839338 | 0.22 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr12_-_16380961 | 0.21 |
ENSDART00000086984
|
hectd2
|
HECT domain containing 2 |
| chr19_+_28256076 | 0.20 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr25_-_7999756 | 0.19 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr11_-_34124319 | 0.16 |
ENSDART00000173275
|
atp13a3
|
ATPase 13A3 |
| chr21_-_38853737 | 0.15 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr2_-_9259283 | 0.13 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr3_+_25825043 | 0.11 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr19_-_10425140 | 0.10 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr23_+_11374168 | 0.08 |
ENSDART00000163083
|
chl1a
|
cell adhesion molecule L1-like a |
| chr23_+_3721042 | 0.07 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr1_-_59422880 | 0.06 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr7_-_41693004 | 0.01 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr18_-_25855263 | 0.00 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.8 | 4.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.6 | 4.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 1.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 2.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.3 | 1.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.0 | GO:0001774 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 1.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.8 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) arachidonate transport(GO:1903963) |
| 0.2 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 3.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.8 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 2.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 2.1 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.8 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 5.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 2.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 2.8 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 1.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 1.2 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.4 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.2 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 2.3 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 3.5 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 2.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 4.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 2.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 3.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 2.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 4.3 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |