Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
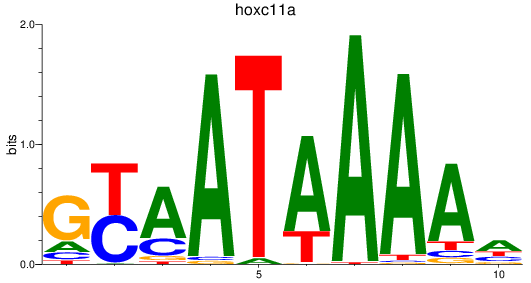
Results for hoxc11a
Z-value: 1.87
Transcription factors associated with hoxc11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11a
|
ENSDARG00000070351 | homeobox C11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11a | dr11_v1_chr23_+_36074798_36074880 | 0.65 | 6.0e-02 | Click! |
Activity profile of hoxc11a motif
Sorted Z-values of hoxc11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_10146542 | 5.33 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr22_-_4644484 | 5.20 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr1_+_44439661 | 4.66 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr10_-_29903165 | 4.15 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr17_+_52822831 | 3.81 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr23_+_39606108 | 3.70 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr12_-_25916530 | 3.68 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr11_+_25257022 | 3.67 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr12_+_27117609 | 3.44 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr11_+_30244356 | 3.42 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr23_+_36052944 | 3.40 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr21_+_5129513 | 3.37 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr23_+_27068225 | 3.31 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr6_-_60147517 | 3.26 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr6_+_52804267 | 3.22 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr25_+_29161609 | 3.12 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr25_+_20089986 | 3.11 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr22_-_14115292 | 2.98 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr6_+_35362225 | 2.90 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr12_+_6041575 | 2.89 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr1_+_46194333 | 2.78 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr13_+_22675802 | 2.78 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr19_-_28789404 | 2.69 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr2_+_2223837 | 2.62 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr5_-_42272517 | 2.59 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr6_+_12853655 | 2.59 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr17_+_52822422 | 2.58 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr6_-_26559921 | 2.53 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr8_+_39619087 | 2.48 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr19_+_19747430 | 2.43 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr9_-_1949915 | 2.36 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr9_-_42696408 | 2.34 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr16_+_32995882 | 2.31 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr20_+_26880668 | 2.31 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr12_-_20373058 | 2.26 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr6_+_40629066 | 2.25 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr3_+_23731109 | 2.16 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr5_-_40510397 | 2.14 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr17_+_52823015 | 2.14 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr20_+_52546186 | 2.11 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_36650222 | 2.06 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr17_-_12389259 | 2.00 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_+_39386982 | 1.99 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr2_+_20406399 | 1.96 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr24_-_6158933 | 1.95 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr21_+_19070921 | 1.95 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr4_+_5741733 | 1.95 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr11_+_6116503 | 1.93 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr23_+_25354856 | 1.91 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr3_+_25154078 | 1.86 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr24_+_5237753 | 1.80 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr11_-_18253111 | 1.77 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr11_+_2198831 | 1.76 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr20_+_6659770 | 1.75 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr19_+_19762183 | 1.74 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr21_+_25236297 | 1.72 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr23_-_12014931 | 1.70 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr3_+_16265924 | 1.70 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr9_-_48370645 | 1.66 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr24_-_8732519 | 1.64 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr7_-_30367650 | 1.63 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr14_+_49135264 | 1.62 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr6_+_58915889 | 1.59 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr7_+_25059845 | 1.58 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr9_+_17306162 | 1.53 |
ENSDART00000075926
|
scel
|
sciellin |
| chr1_-_35924495 | 1.48 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr18_-_15373620 | 1.46 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr18_+_48446704 | 1.43 |
ENSDART00000134817
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr7_+_36041509 | 1.42 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr8_-_34052019 | 1.40 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr15_-_34567370 | 1.39 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr10_+_26612321 | 1.36 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr8_+_19668654 | 1.34 |
ENSDART00000091436
|
foxe3
|
forkhead box E3 |
| chr20_+_43925266 | 1.34 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr16_-_26676685 | 1.34 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr20_-_25709247 | 1.33 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr9_+_44431174 | 1.30 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr23_-_7799184 | 1.28 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr9_+_34425736 | 1.27 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr4_-_5652030 | 1.27 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr1_+_36437585 | 1.25 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr24_+_9412450 | 1.25 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr11_+_42556395 | 1.23 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr21_-_25756119 | 1.23 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr8_+_14381272 | 1.19 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr7_-_26844064 | 1.18 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr18_-_15559817 | 1.18 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr11_-_21586157 | 1.18 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_-_43666166 | 1.16 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr11_+_21053488 | 1.15 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr19_-_10243148 | 1.14 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr17_-_17764801 | 1.11 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr7_+_30875273 | 1.11 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr1_+_5402476 | 1.09 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr14_-_12837432 | 1.06 |
ENSDART00000178444
|
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr5_+_11290851 | 1.06 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr1_-_22512063 | 1.05 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr25_-_26758253 | 1.05 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr14_-_12837052 | 1.04 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr5_+_24245682 | 1.04 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr1_+_27868294 | 1.02 |
ENSDART00000165332
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr16_+_10918252 | 1.02 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr24_+_39105051 | 0.99 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr1_-_39976492 | 0.97 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr9_-_11550711 | 0.96 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr22_+_5176255 | 0.95 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr16_+_23087326 | 0.95 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr7_+_61480296 | 0.95 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr15_-_19772372 | 0.94 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr9_+_44430974 | 0.93 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr3_+_24134418 | 0.93 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr11_-_6265574 | 0.93 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr9_-_16853462 | 0.92 |
ENSDART00000160273
|
CT573248.2
|
|
| chr17_-_30975707 | 0.92 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr19_+_19761966 | 0.90 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr9_+_917060 | 0.89 |
ENSDART00000082390
|
tmem37
|
transmembrane protein 37 |
| chr23_+_18722715 | 0.87 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr20_+_30490682 | 0.86 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr9_-_41784799 | 0.86 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr10_-_10864331 | 0.86 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr6_-_40697585 | 0.86 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr2_+_52847049 | 0.86 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr8_-_52413032 | 0.84 |
ENSDART00000183039
|
CABZ01070469.1
|
|
| chr2_-_27329667 | 0.84 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr17_-_19019635 | 0.84 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_50203682 | 0.82 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr25_+_37126921 | 0.82 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr8_-_25327809 | 0.79 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr11_-_15090564 | 0.79 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr23_+_28648864 | 0.79 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr11_-_6188413 | 0.79 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr10_+_2587234 | 0.77 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr20_-_47731768 | 0.76 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr23_+_23918421 | 0.76 |
ENSDART00000046951
|
ptpn11b
|
protein tyrosine phosphatase, non-receptor type 11, b |
| chr17_-_18888959 | 0.75 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr3_+_62161184 | 0.75 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr3_-_19200571 | 0.75 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr16_+_44768361 | 0.74 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr5_+_37785152 | 0.73 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr2_+_5927255 | 0.73 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr20_+_20726231 | 0.73 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr9_+_41080029 | 0.70 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr20_-_27733683 | 0.70 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr11_-_12998400 | 0.68 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr10_+_16069987 | 0.68 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr15_-_17869115 | 0.67 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr20_-_4883673 | 0.67 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr2_-_30659222 | 0.65 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr24_+_5840258 | 0.64 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr24_+_25259154 | 0.64 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr11_+_14284866 | 0.63 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr6_+_23931236 | 0.62 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr10_-_34772211 | 0.62 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr10_-_34889053 | 0.61 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr5_-_63218919 | 0.60 |
ENSDART00000149979
|
tecta
|
tectorin alpha |
| chr3_+_24190207 | 0.59 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr13_+_29519115 | 0.57 |
ENSDART00000086711
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr13_+_36585399 | 0.56 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr4_-_56898328 | 0.56 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr11_-_29737088 | 0.55 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr4_-_71177920 | 0.55 |
ENSDART00000158287
|
si:dkey-193i10.1
|
si:dkey-193i10.1 |
| chr1_+_27153859 | 0.55 |
ENSDART00000180184
|
bnc2
|
basonuclin 2 |
| chr1_+_961607 | 0.53 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr9_+_54290896 | 0.53 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr2_+_45068366 | 0.52 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr16_+_13883872 | 0.50 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr7_-_26457208 | 0.48 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr7_-_29341233 | 0.48 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr2_-_27329214 | 0.48 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr6_-_6448519 | 0.47 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr11_+_41243325 | 0.47 |
ENSDART00000170657
|
pax7a
|
paired box 7a |
| chr10_-_25591194 | 0.46 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr23_-_27571667 | 0.45 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr2_-_17235891 | 0.44 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr8_+_30709685 | 0.42 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr1_-_19215336 | 0.41 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr7_+_18075504 | 0.39 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr23_+_7379728 | 0.38 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr11_-_15090118 | 0.38 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr2_-_9607879 | 0.37 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr19_+_1003332 | 0.37 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr22_+_31821815 | 0.37 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr4_+_43522945 | 0.36 |
ENSDART00000183921
ENSDART00000181832 |
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr8_-_19280856 | 0.35 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr12_-_46959990 | 0.35 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr2_+_19785540 | 0.34 |
ENSDART00000149789
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr20_+_6543674 | 0.34 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr20_+_27428505 | 0.34 |
ENSDART00000002641
|
kif26aa
|
kinesin family member 26Aa |
| chr15_+_14856307 | 0.34 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr24_+_4434117 | 0.32 |
ENSDART00000184349
|
GJD4
|
gap junction protein delta 4 |
| chr1_+_44826593 | 0.31 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr5_+_31946973 | 0.31 |
ENSDART00000189876
ENSDART00000163366 |
ungb
|
uracil DNA glycosylase b |
| chr19_+_42469058 | 0.30 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr18_-_14941840 | 0.30 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_-_26610814 | 0.29 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr17_-_47090440 | 0.29 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr19_+_7810028 | 0.29 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr1_-_9228007 | 0.28 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr21_-_20328375 | 0.28 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr17_+_31221761 | 0.28 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.7 | 2.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 3.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 1.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 1.3 | GO:1902746 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 1.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 3.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 1.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.4 | 3.6 | GO:0006833 | water transport(GO:0006833) |
| 0.4 | 1.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 2.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 1.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 1.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 0.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 2.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 1.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 1.7 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.2 | 2.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 3.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 3.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 5.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 1.6 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.4 | GO:0061687 | DNA damage response, detection of DNA damage(GO:0042769) detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.9 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.5 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 2.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 6.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 0.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 3.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.5 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.1 | 1.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 3.6 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 1.0 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 4.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 3.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 2.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.2 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.7 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 5.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.5 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 4.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.3 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 2.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 3.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.7 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 0.9 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 5.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.0 | 3.0 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.7 | 2.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 5.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.6 | 1.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.5 | 3.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 1.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 3.3 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 1.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.2 | 1.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 5.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.6 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 2.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 8.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 13.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 9.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 3.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 28.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 3.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |