Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
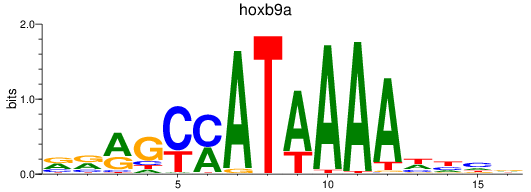
Results for hoxb9a
Z-value: 0.92
Transcription factors associated with hoxb9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb9a
|
ENSDARG00000056023 | homeobox B9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb9a | dr11_v1_chr3_+_23677351_23677351 | 0.50 | 1.7e-01 | Click! |
Activity profile of hoxb9a motif
Sorted Z-values of hoxb9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_7379728 | 1.96 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr23_+_39606108 | 1.41 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr6_+_35362225 | 1.32 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr8_+_4697764 | 1.18 |
ENSDART00000064197
|
cldn5a
|
claudin 5a |
| chr5_-_40510397 | 1.18 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr2_-_40135942 | 0.98 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr2_+_52847049 | 0.97 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr24_-_17029374 | 0.95 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr10_+_506538 | 0.95 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr13_+_22295905 | 0.90 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr22_-_4644484 | 0.87 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr13_+_43400443 | 0.79 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr17_-_44249720 | 0.75 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr20_+_52546186 | 0.69 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr4_-_56898328 | 0.66 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr13_-_49802194 | 0.63 |
ENSDART00000148722
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr13_+_1182257 | 0.61 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr15_-_21014015 | 0.59 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr21_-_11970199 | 0.58 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr7_+_36041509 | 0.57 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr10_-_3332362 | 0.57 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr15_-_19772372 | 0.57 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr2_-_30668580 | 0.56 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr25_-_31739309 | 0.56 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr2_+_15586632 | 0.55 |
ENSDART00000164903
|
BX510342.1
|
|
| chr9_-_3400727 | 0.55 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr24_+_39105051 | 0.55 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr8_+_45004997 | 0.54 |
ENSDART00000159779
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr9_+_32860345 | 0.54 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr3_-_49554912 | 0.53 |
ENSDART00000159392
|
CR847534.1
|
|
| chr21_+_25236297 | 0.52 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr10_-_22127942 | 0.51 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr24_-_6345647 | 0.51 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr4_-_59709185 | 0.51 |
ENSDART00000136825
|
si:dkey-149m13.5
|
si:dkey-149m13.5 |
| chr22_+_21255860 | 0.51 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr2_-_44283554 | 0.50 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr4_+_45504938 | 0.48 |
ENSDART00000145958
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr2_-_3158919 | 0.48 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr8_+_45004666 | 0.46 |
ENSDART00000145348
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr21_-_39931285 | 0.46 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr6_+_43015916 | 0.45 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr7_+_61480296 | 0.45 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr16_-_55028740 | 0.44 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr17_+_33375469 | 0.44 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr3_+_6469754 | 0.43 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr25_-_29134000 | 0.43 |
ENSDART00000172027
ENSDART00000190447 |
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr4_+_42556555 | 0.42 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr3_-_31804481 | 0.40 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr4_+_59711338 | 0.38 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr2_+_13462305 | 0.38 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr8_-_18262149 | 0.38 |
ENSDART00000143000
|
rnf220b
|
ring finger protein 220b |
| chr5_-_67750907 | 0.37 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr6_+_39536596 | 0.37 |
ENSDART00000185742
|
CU467856.3
|
|
| chr2_+_19785540 | 0.36 |
ENSDART00000149789
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr9_+_32859967 | 0.35 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr4_-_55641422 | 0.34 |
ENSDART00000165178
|
znf1074
|
zinc finger protein 1074 |
| chr16_-_43679611 | 0.34 |
ENSDART00000123585
|
LO017721.1
|
|
| chr23_+_36122058 | 0.34 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr10_+_16069987 | 0.32 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr4_+_37406676 | 0.32 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr4_-_59159690 | 0.32 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr7_-_2193583 | 0.31 |
ENSDART00000171574
|
CABZ01007816.1
|
|
| chr9_-_48370645 | 0.30 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr14_+_22447662 | 0.30 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr14_+_14841685 | 0.29 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr13_-_33362191 | 0.29 |
ENSDART00000100514
|
zgc:172120
|
zgc:172120 |
| chr4_-_50788075 | 0.29 |
ENSDART00000150302
|
znf1045
|
zinc finger protein 1045 |
| chr14_+_30795559 | 0.28 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr19_+_4916233 | 0.28 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr10_-_42923385 | 0.28 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr4_-_71108793 | 0.27 |
ENSDART00000189856
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr11_-_18253111 | 0.26 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr19_+_19747430 | 0.26 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr12_-_33817114 | 0.26 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr3_-_32958505 | 0.24 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr25_-_12923482 | 0.24 |
ENSDART00000161754
|
CR450808.1
|
|
| chr16_+_38940758 | 0.24 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr24_-_3426620 | 0.24 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr4_-_64142389 | 0.24 |
ENSDART00000172126
|
BX914205.3
|
|
| chr6_-_54107269 | 0.23 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr14_-_41388178 | 0.23 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr8_-_3312384 | 0.23 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr8_-_7847921 | 0.23 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr8_-_38159805 | 0.23 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr12_-_20373058 | 0.23 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr4_-_73299225 | 0.23 |
ENSDART00000174178
ENSDART00000174198 |
si:cabz01021430.2
|
si:cabz01021430.2 |
| chr25_+_20089986 | 0.22 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr13_+_36585399 | 0.22 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr25_+_32755485 | 0.21 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr22_+_15343953 | 0.21 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr8_-_50888806 | 0.20 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr12_+_27022517 | 0.20 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr8_+_22490069 | 0.20 |
ENSDART00000135721
|
si:ch211-261n11.8
|
si:ch211-261n11.8 |
| chr4_-_61920018 | 0.20 |
ENSDART00000164832
|
znf1056
|
zinc finger protein 1056 |
| chr14_+_15495088 | 0.20 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr3_-_34624745 | 0.19 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr11_+_12175162 | 0.19 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr6_+_13083146 | 0.18 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr3_+_24190207 | 0.18 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr15_+_32405959 | 0.18 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr20_-_47270519 | 0.18 |
ENSDART00000153329
|
si:dkeyp-104f11.8
|
si:dkeyp-104f11.8 |
| chr14_-_9056066 | 0.18 |
ENSDART00000139669
ENSDART00000138758 ENSDART00000041099 |
sybl1
|
synaptobrevin-like 1 |
| chr5_-_30984271 | 0.17 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr4_-_30712588 | 0.17 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr25_-_13842618 | 0.17 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr5_+_24087035 | 0.17 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr25_-_31898552 | 0.17 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr4_-_73190246 | 0.17 |
ENSDART00000170842
|
LO018260.1
|
|
| chr17_-_43666166 | 0.17 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr23_+_18722915 | 0.17 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr23_+_18722715 | 0.17 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr15_+_21254800 | 0.17 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr15_+_11381532 | 0.16 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr22_+_6321038 | 0.15 |
ENSDART00000179961
|
si:rp71-1i20.1
|
si:rp71-1i20.1 |
| chr2_+_19578079 | 0.15 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr17_-_23727978 | 0.15 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr1_+_45663727 | 0.15 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr7_+_33152723 | 0.14 |
ENSDART00000132658
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr1_+_56463494 | 0.14 |
ENSDART00000097964
|
zgc:171452
|
zgc:171452 |
| chr12_+_10053852 | 0.13 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr1_-_56556326 | 0.13 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr10_-_40589641 | 0.13 |
ENSDART00000140228
ENSDART00000154850 |
taar16b
|
trace amine associated receptor 16b |
| chr14_-_1958994 | 0.12 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr2_+_5927255 | 0.11 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr4_+_33574463 | 0.11 |
ENSDART00000150255
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr14_+_5861435 | 0.10 |
ENSDART00000041279
ENSDART00000147341 |
tubb4b
|
tubulin, beta 4B class IVb |
| chr12_+_18681477 | 0.10 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr10_+_40700311 | 0.10 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr2_-_40196547 | 0.09 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr11_-_38533505 | 0.09 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr14_+_4276394 | 0.09 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr11_-_25538341 | 0.09 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr24_-_14712427 | 0.08 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr4_+_39368978 | 0.08 |
ENSDART00000160640
|
si:dkey-261o4.1
|
si:dkey-261o4.1 |
| chr4_-_77300601 | 0.08 |
ENSDART00000163015
ENSDART00000174015 |
slco1f2
|
solute carrier organic anion transporter family, member 1F2 |
| chr10_-_24371312 | 0.07 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr20_-_40720458 | 0.07 |
ENSDART00000153151
ENSDART00000061261 ENSDART00000138569 |
cx43
|
connexin 43 |
| chr4_-_46915962 | 0.07 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr21_+_11969603 | 0.06 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr10_+_40660772 | 0.06 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr10_+_40768203 | 0.06 |
ENSDART00000171994
ENSDART00000140343 |
taar19d
|
trace amine associated receptor 19d |
| chr24_-_21819010 | 0.05 |
ENSDART00000091096
|
CR352265.1
|
|
| chr5_+_72194444 | 0.05 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr24_+_29352039 | 0.05 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr16_-_7793457 | 0.05 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr1_+_44439661 | 0.04 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr2_-_32637592 | 0.04 |
ENSDART00000136353
|
si:dkeyp-73d8.8
|
si:dkeyp-73d8.8 |
| chr23_+_20931030 | 0.04 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr19_-_47571456 | 0.04 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr6_+_41956355 | 0.04 |
ENSDART00000176856
|
oxtr
|
oxytocin receptor |
| chr22_-_10605045 | 0.03 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr3_+_36646054 | 0.03 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr25_+_7299488 | 0.03 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr21_-_2415808 | 0.03 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr7_+_29512673 | 0.03 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr8_-_6897619 | 0.03 |
ENSDART00000133606
|
si:ch211-255g12.8
|
si:ch211-255g12.8 |
| chr20_+_33532296 | 0.03 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr20_-_29499363 | 0.03 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr22_+_10606573 | 0.03 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr7_+_30875273 | 0.03 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr15_+_32643873 | 0.02 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr22_+_8612462 | 0.02 |
ENSDART00000114586
|
CR450686.1
|
|
| chr24_+_1042594 | 0.02 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr3_-_25490970 | 0.01 |
ENSDART00000184651
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr25_+_4750972 | 0.01 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr17_-_21057617 | 0.01 |
ENSDART00000148095
ENSDART00000048853 |
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr3_-_53091946 | 0.01 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr13_+_27329556 | 0.01 |
ENSDART00000140085
|
mb21d1
|
Mab-21 domain containing 1 |
| chr11_+_42585138 | 0.01 |
ENSDART00000019008
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr25_-_19219807 | 0.00 |
ENSDART00000183577
|
acanb
|
aggrecan b |
| chr6_-_6345991 | 0.00 |
ENSDART00000174445
|
ccdc88aa
|
coiled-coil domain containing 88Aa |
| chr8_-_52413032 | 0.00 |
ENSDART00000183039
|
CABZ01070469.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.3 | 0.9 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 2.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.0 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.2 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.2 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 1.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.0 | GO:0007567 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.6 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.2 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 1.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |