Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
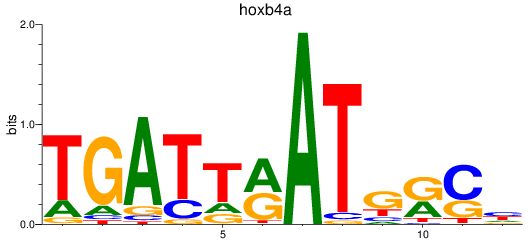
Results for hoxb4a
Z-value: 0.50
Transcription factors associated with hoxb4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb4a
|
ENSDARG00000013533 | homeobox B4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb4a | dr11_v1_chr3_+_23721808_23721808 | -0.96 | 3.6e-05 | Click! |
Activity profile of hoxb4a motif
Sorted Z-values of hoxb4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_57473980 | 0.83 |
ENSDART00000149353
ENSDART00000150034 |
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr24_+_26402110 | 0.83 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr4_-_17257435 | 0.79 |
ENSDART00000131973
|
lrmp
|
lymphoid-restricted membrane protein |
| chr17_+_19626479 | 0.77 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr11_-_13341051 | 0.74 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr23_+_27756984 | 0.66 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr13_-_33207367 | 0.65 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr15_+_25489406 | 0.64 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr11_-_13341483 | 0.62 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr25_+_5604512 | 0.62 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr25_+_5972690 | 0.62 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr5_-_18446483 | 0.61 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr12_-_20616160 | 0.59 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr6_+_18367388 | 0.56 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr5_-_64103863 | 0.54 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr18_-_12957451 | 0.53 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr13_-_22961605 | 0.52 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr25_-_29072162 | 0.51 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_+_19687217 | 0.51 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr2_+_44972720 | 0.46 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr7_-_19369002 | 0.46 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr4_-_13255700 | 0.46 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr15_-_15968883 | 0.42 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr11_+_18205766 | 0.41 |
ENSDART00000175559
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr21_+_20901505 | 0.39 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr19_+_33139164 | 0.38 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr9_+_45605410 | 0.35 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr2_+_23731194 | 0.35 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr23_+_384850 | 0.35 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr16_-_32672883 | 0.29 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr16_-_18960613 | 0.29 |
ENSDART00000183197
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr5_+_20112032 | 0.29 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr18_-_42172101 | 0.29 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr7_+_13988075 | 0.26 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr9_-_3653259 | 0.26 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr2_-_36818132 | 0.25 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr9_-_15789526 | 0.25 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr13_+_18321140 | 0.24 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr23_-_3721444 | 0.22 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr18_+_6126506 | 0.22 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr17_-_114121 | 0.22 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr19_+_20177208 | 0.20 |
ENSDART00000166807
|
tra2a
|
transformer 2 alpha homolog |
| chr8_+_21406769 | 0.18 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr4_+_43408004 | 0.17 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr1_+_1712140 | 0.17 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr6_-_43047774 | 0.16 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr18_-_25401002 | 0.14 |
ENSDART00000055567
|
gnrhr4
|
gonadotropin releasing hormone receptor 4 |
| chr11_+_13223625 | 0.12 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr2_+_37480669 | 0.11 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr24_+_32472155 | 0.11 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr5_+_32924669 | 0.11 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr1_+_11881559 | 0.10 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr3_-_51109286 | 0.09 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr25_-_4482449 | 0.08 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr1_-_11372456 | 0.07 |
ENSDART00000144164
ENSDART00000141238 |
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr23_-_33038423 | 0.07 |
ENSDART00000180539
|
plxna2
|
plexin A2 |
| chr14_-_30876299 | 0.06 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr4_-_2637689 | 0.06 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr14_-_30876708 | 0.06 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr17_-_8727699 | 0.06 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr20_-_32188897 | 0.05 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr10_-_15910974 | 0.04 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr6_+_34870374 | 0.04 |
ENSDART00000149356
|
il23r
|
interleukin 23 receptor |
| chr19_-_10425140 | 0.04 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr8_-_912821 | 0.02 |
ENSDART00000082296
|
ptger4a
|
prostaglandin E receptor 4 (subtype EP4) a |
| chr15_+_17251191 | 0.02 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr7_+_27317174 | 0.01 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr23_+_3721042 | 0.01 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr19_+_28256076 | 0.01 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.7 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.5 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |