Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
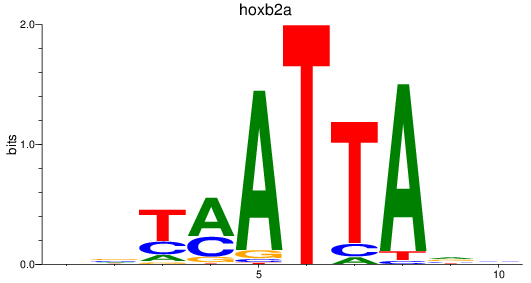
Results for hoxb2a
Z-value: 1.21
Transcription factors associated with hoxb2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb2a
|
ENSDARG00000000175 | homeobox B2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb2a | dr11_v1_chr3_+_23752150_23752150 | -0.82 | 6.8e-03 | Click! |
Activity profile of hoxb2a motif
Sorted Z-values of hoxb2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 5.00 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr19_-_18313303 | 3.12 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr18_+_20560616 | 3.05 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr24_+_1023839 | 2.97 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr20_-_23426339 | 2.82 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr10_-_44560165 | 2.62 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr10_-_21362071 | 2.42 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 2.38 |
ENSDART00000189789
|
avd
|
avidin |
| chr24_-_10014512 | 2.21 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr5_-_19006290 | 2.02 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr10_-_25217347 | 1.99 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr6_+_28208973 | 1.96 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr4_-_77125693 | 1.94 |
ENSDART00000174256
|
CU467646.3
|
|
| chr1_-_18811517 | 1.74 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr11_-_1550709 | 1.73 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr14_-_33481428 | 1.71 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr1_-_55248496 | 1.71 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr16_-_42056137 | 1.59 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr10_+_6884627 | 1.57 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr20_-_38787047 | 1.57 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr2_+_6253246 | 1.54 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_-_38787341 | 1.51 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr9_-_38021889 | 1.47 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr8_+_41037541 | 1.45 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr19_+_31585917 | 1.44 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr21_-_14174786 | 1.42 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr2_-_57076687 | 1.36 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr5_-_26247973 | 1.34 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr6_-_43283122 | 1.34 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr5_-_30074332 | 1.34 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr3_+_26244353 | 1.33 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr9_+_29548195 | 1.33 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr5_-_16734060 | 1.31 |
ENSDART00000035859
ENSDART00000190457 ENSDART00000179244 |
atad1a
|
ATPase family, AAA domain containing 1a |
| chr15_-_16177603 | 1.31 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr14_+_24845941 | 1.30 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr17_+_16046132 | 1.28 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_+_31864921 | 1.26 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr14_-_8940499 | 1.25 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr22_-_22337382 | 1.25 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr10_+_6884123 | 1.24 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr6_-_2134581 | 1.23 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr22_+_1170294 | 1.21 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr8_-_1838315 | 1.21 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr25_-_37262220 | 1.19 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr2_+_41526904 | 1.19 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr5_+_40837539 | 1.18 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr20_-_48898371 | 1.18 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr14_-_33945692 | 1.16 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr23_-_35790235 | 1.12 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr22_+_14836040 | 1.10 |
ENSDART00000180951
|
gtpbp1l
|
GTP binding protein 1, like |
| chr5_-_11809710 | 1.10 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr22_+_4488454 | 1.09 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr17_-_40956035 | 1.09 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr23_+_44236281 | 1.08 |
ENSDART00000149842
|
MEPCE
|
si:ch1073-157b13.1 |
| chr10_+_15255198 | 1.08 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr16_+_47207691 | 1.08 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr20_+_34671386 | 1.07 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr5_+_29794058 | 1.06 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr16_+_29509133 | 1.06 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr6_-_41138854 | 1.06 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr6_+_21001264 | 1.06 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr3_-_26244256 | 1.05 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr16_+_9400661 | 1.04 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr10_-_35257458 | 1.04 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr11_+_35171406 | 1.04 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr16_-_42965192 | 1.02 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr7_+_34592526 | 1.01 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr6_+_40922572 | 0.99 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr15_-_44601331 | 0.99 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr19_-_30510259 | 0.99 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr23_+_42254960 | 0.99 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr22_-_17671348 | 0.98 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr25_-_21031007 | 0.97 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr1_-_23308225 | 0.97 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr17_+_16046314 | 0.96 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_+_18807006 | 0.95 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr17_+_49500820 | 0.94 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr22_-_10440688 | 0.93 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr5_+_63302660 | 0.92 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr21_+_1647990 | 0.92 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr16_+_25011994 | 0.91 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr25_+_35891342 | 0.90 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr21_-_32060993 | 0.90 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr21_-_19918286 | 0.89 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr21_-_31013817 | 0.89 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr23_-_31913069 | 0.89 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_+_26722904 | 0.89 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr12_-_34258384 | 0.88 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr15_-_17169935 | 0.88 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr5_-_9625459 | 0.88 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr18_+_18000887 | 0.88 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr15_-_43978141 | 0.87 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr11_-_45152702 | 0.87 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr1_-_55068941 | 0.86 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr15_-_34930727 | 0.86 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr10_-_32494499 | 0.86 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr19_+_2631565 | 0.86 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr24_+_39518774 | 0.85 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr19_+_40069524 | 0.84 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr6_-_40922971 | 0.84 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr3_-_16719244 | 0.83 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr1_+_40613297 | 0.83 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr8_+_8238727 | 0.83 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr3_+_22035863 | 0.82 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr12_+_23912074 | 0.81 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr12_+_30367371 | 0.81 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr4_+_11723852 | 0.80 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_+_16151368 | 0.79 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_-_32494304 | 0.78 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr4_-_72296520 | 0.78 |
ENSDART00000182638
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr8_+_11425048 | 0.77 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr23_-_32100106 | 0.77 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr13_+_1015749 | 0.76 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr21_-_30994577 | 0.75 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr5_+_27137473 | 0.75 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr11_-_34219211 | 0.75 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr6_-_3982783 | 0.75 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr10_+_1849874 | 0.75 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr5_+_6954162 | 0.74 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr16_+_28994709 | 0.73 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr14_-_38889311 | 0.73 |
ENSDART00000186978
|
zgc:101583
|
zgc:101583 |
| chr9_+_54695981 | 0.73 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr15_-_34408777 | 0.73 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr19_+_42227400 | 0.72 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr3_+_28860283 | 0.72 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr7_+_24573721 | 0.72 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr17_-_31611692 | 0.72 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr18_+_18000695 | 0.71 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr18_-_20458840 | 0.71 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr8_+_25145464 | 0.71 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr17_-_41798856 | 0.71 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr4_+_65127317 | 0.71 |
ENSDART00000166475
|
znf1126
|
zinc finger protein 1126 |
| chr12_-_24060358 | 0.70 |
ENSDART00000050831
|
chac2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr21_-_3853204 | 0.68 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr18_+_44532199 | 0.68 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr7_+_6915609 | 0.68 |
ENSDART00000159213
|
ccs
|
copper chaperone for superoxide dismutase |
| chr20_+_46513651 | 0.68 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr10_-_41302841 | 0.67 |
ENSDART00000020297
ENSDART00000160174 ENSDART00000183850 ENSDART00000169493 |
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr9_+_17983463 | 0.67 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr21_+_45502773 | 0.67 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr10_+_518546 | 0.67 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr16_-_17347727 | 0.67 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr20_+_36629173 | 0.66 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr10_+_35554219 | 0.65 |
ENSDART00000077373
|
zdhhc20a
|
zinc finger, DHHC-type containing 20a |
| chr24_-_2450597 | 0.65 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr1_+_21937201 | 0.64 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr7_+_17816006 | 0.64 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr4_-_65037243 | 0.64 |
ENSDART00000170059
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr5_+_66433287 | 0.64 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr12_-_3077395 | 0.64 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr12_-_18577983 | 0.63 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr20_-_9428021 | 0.63 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr20_-_45060241 | 0.63 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr23_-_900795 | 0.62 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr2_+_41524238 | 0.62 |
ENSDART00000122860
ENSDART00000017977 |
acvr1l
|
activin A receptor, type 1 like |
| chr2_-_55298075 | 0.62 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr18_-_43884044 | 0.62 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr7_-_64589920 | 0.62 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr15_+_17345609 | 0.62 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr21_-_25801956 | 0.62 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr5_-_26765188 | 0.61 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr7_-_7764287 | 0.60 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr17_+_8799451 | 0.60 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr24_-_34680956 | 0.60 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_+_23515966 | 0.59 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr12_+_16087077 | 0.59 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr17_-_48944465 | 0.59 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr11_-_28050559 | 0.59 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr7_-_17816175 | 0.59 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr1_-_47071979 | 0.59 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr23_+_32028574 | 0.59 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr11_+_34523132 | 0.58 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr12_+_30367079 | 0.58 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr5_+_60934534 | 0.58 |
ENSDART00000065025
|
rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr22_-_6362442 | 0.57 |
ENSDART00000149907
ENSDART00000106104 |
zgc:113298
|
zgc:113298 |
| chr7_+_17816470 | 0.57 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr23_-_31913231 | 0.57 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_-_12464412 | 0.56 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr3_+_16663373 | 0.55 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr18_-_18875308 | 0.55 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr7_+_67749251 | 0.55 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr21_+_34088377 | 0.55 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr23_-_14990865 | 0.55 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr19_-_5805923 | 0.55 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr1_-_26444075 | 0.55 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr15_+_1534644 | 0.54 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr9_-_5318873 | 0.54 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr20_+_22067337 | 0.54 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr20_+_38458084 | 0.54 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr8_+_7801060 | 0.54 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr9_-_7287128 | 0.53 |
ENSDART00000176281
ENSDART00000065803 |
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr21_+_4313039 | 0.52 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr11_-_35171768 | 0.52 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr18_+_6558338 | 0.52 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr10_-_33297864 | 0.52 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr5_-_25733745 | 0.51 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr19_+_12406583 | 0.51 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr12_-_18393408 | 0.51 |
ENSDART00000159674
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr5_-_8907819 | 0.50 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr15_+_31344472 | 0.50 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.0 | 3.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 | 1.8 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.4 | 1.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 1.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 1.1 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.3 | 3.1 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.3 | 2.8 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.3 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 1.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.7 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 3.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 2.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 1.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.6 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.3 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.4 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.9 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.4 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.5 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.9 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.6 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.7 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.7 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 2.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 1.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.7 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.5 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.5 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 1.5 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.0 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.9 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.6 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 2.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 1.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.6 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 1.4 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0021576 | hindbrain formation(GO:0021576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 1.8 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.3 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.4 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 4.5 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.7 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 5.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 3.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.5 | 1.4 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 1.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 0.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.3 | 1.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 0.7 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.2 | 3.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 2.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 1.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 1.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.7 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.7 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.3 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.3 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 5.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 3.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 5.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 4.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |