Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
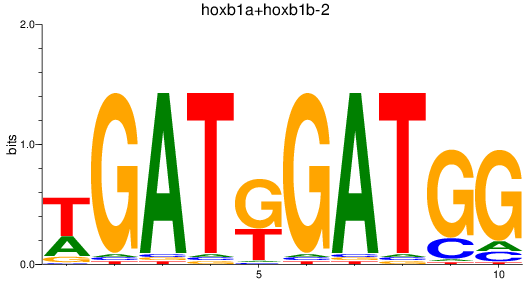
Results for hoxb1a+hoxb1b-2
Z-value: 0.90
Transcription factors associated with hoxb1a+hoxb1b-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb1a
|
ENSDARG00000008174 | homeobox B1a |
|
hoxb1b-2
|
ENSDARG00000112663 | homeobox B1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb1a | dr11_v1_chr3_+_23768898_23768898 | 0.26 | 5.0e-01 | Click! |
Activity profile of hoxb1a+hoxb1b-2 motif
Sorted Z-values of hoxb1a+hoxb1b-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_29952719 | 2.02 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr16_+_1383914 | 1.97 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr6_+_29791164 | 1.73 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr25_+_20215964 | 1.62 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr14_-_36378494 | 1.52 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr25_+_20216159 | 1.46 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr1_-_38709551 | 1.42 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr21_-_28920245 | 1.34 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr14_+_34547554 | 1.31 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr2_+_55984788 | 1.22 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr12_-_25916530 | 1.12 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr7_-_13381129 | 1.06 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr9_+_22003942 | 0.96 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr1_-_20911297 | 0.93 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr3_+_23768898 | 0.93 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr2_+_27010439 | 0.91 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr8_+_6954984 | 0.89 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr20_-_35841570 | 0.89 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr7_+_31879649 | 0.85 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_-_31188715 | 0.85 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr14_-_7888748 | 0.83 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr8_+_44703864 | 0.77 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr16_+_46111849 | 0.74 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr1_-_8428736 | 0.72 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr3_-_57744323 | 0.70 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr22_-_600016 | 0.68 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr10_+_439692 | 0.68 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr22_+_3223489 | 0.66 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr4_+_21129752 | 0.62 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr12_-_10220036 | 0.60 |
ENSDART00000134619
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr23_+_36178104 | 0.60 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr8_-_39903803 | 0.59 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr10_+_1396940 | 0.58 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr25_+_7461624 | 0.56 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr11_+_37201483 | 0.56 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr24_-_20016817 | 0.56 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr18_+_40354998 | 0.55 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr2_+_2223837 | 0.55 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr24_+_34113424 | 0.54 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr23_-_11870962 | 0.51 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr7_+_25036188 | 0.51 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr10_-_44305180 | 0.50 |
ENSDART00000187552
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr18_+_24922125 | 0.48 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr6_-_13401133 | 0.47 |
ENSDART00000151206
|
fmnl2b
|
formin-like 2b |
| chr25_-_13842618 | 0.46 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr22_-_16997475 | 0.46 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr7_-_52842007 | 0.44 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr20_-_25369767 | 0.44 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr10_-_27049170 | 0.42 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr16_+_10422836 | 0.41 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr16_-_37964325 | 0.38 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr4_+_26496489 | 0.38 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr17_+_51764310 | 0.38 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr4_-_8014463 | 0.38 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr13_-_31829786 | 0.37 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr1_+_26071869 | 0.36 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr23_+_20408227 | 0.36 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr7_-_33350082 | 0.36 |
ENSDART00000008785
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr4_+_4272622 | 0.36 |
ENSDART00000164810
|
ano2
|
anoctamin 2 |
| chr18_+_28102620 | 0.34 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr2_-_53525896 | 0.34 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr22_-_16997246 | 0.32 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr21_-_31290582 | 0.32 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr5_-_22544151 | 0.31 |
ENSDART00000006474
|
glra4b
|
glycine receptor, alpha 4b |
| chr3_+_15805917 | 0.31 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr11_-_25213651 | 0.31 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr20_-_49650293 | 0.31 |
ENSDART00000151239
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr17_-_20897250 | 0.30 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr8_-_7440364 | 0.29 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr11_-_27702778 | 0.28 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr3_-_35800221 | 0.28 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr5_-_66028371 | 0.27 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr5_-_52921532 | 0.25 |
ENSDART00000160566
|
CU695117.1
|
|
| chr22_-_13544244 | 0.23 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr24_+_28528000 | 0.23 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr8_+_6410933 | 0.23 |
ENSDART00000168789
|
lrrtm4l2
|
leucine rich repeat transmembrane neuronal 4 like 2 |
| chr10_-_25591194 | 0.23 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr16_-_1502699 | 0.22 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr19_+_816208 | 0.22 |
ENSDART00000093304
|
nrm
|
nurim |
| chr8_-_5220125 | 0.21 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr18_+_26899316 | 0.20 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr1_+_34181581 | 0.19 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr5_-_52277643 | 0.19 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr3_-_18675688 | 0.19 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr7_+_16835218 | 0.19 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr12_+_22580579 | 0.18 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr8_-_44015210 | 0.17 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr23_+_642001 | 0.17 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr1_-_681116 | 0.17 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr16_-_17175731 | 0.16 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr17_-_20394566 | 0.14 |
ENSDART00000154667
|
sorcs3b
|
sortilin related VPS10 domain containing receptor 3b |
| chr14_-_26377044 | 0.14 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr14_+_5383060 | 0.13 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr24_+_32472155 | 0.11 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr1_-_20826023 | 0.11 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr7_+_36898850 | 0.11 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr2_-_45630823 | 0.10 |
ENSDART00000183553
|
CR450720.1
|
|
| chr17_+_40684 | 0.10 |
ENSDART00000164231
|
CABZ01111555.1
|
|
| chr17_-_20897407 | 0.09 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr9_+_22351443 | 0.07 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr10_-_31175744 | 0.06 |
ENSDART00000191728
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr13_-_51065048 | 0.05 |
ENSDART00000168085
|
CABZ01078663.1
|
|
| chr19_+_28256076 | 0.04 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr23_+_642395 | 0.04 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr6_+_3640381 | 0.04 |
ENSDART00000172078
|
col28a2b
|
collagen, type XXVIII, alpha 2b |
| chr5_+_35398745 | 0.03 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr18_+_41542542 | 0.03 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr17_-_19626357 | 0.03 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr16_+_25196572 | 0.02 |
ENSDART00000141956
|
tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr16_-_20932896 | 0.02 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr2_-_26563978 | 0.02 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr10_+_7564106 | 0.01 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr16_-_1503023 | 0.01 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr1_+_26626824 | 0.00 |
ENSDART00000158193
|
coro2a
|
coronin, actin binding protein, 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb1a+hoxb1b-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 0.6 | GO:0090189 | regulation of neurotransmitter uptake(GO:0051580) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 0.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.9 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 0.8 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 3.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.9 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.4 | GO:0097009 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.0 | 2.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.9 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 3.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 3.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 1.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 1.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 2.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.4 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |