Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
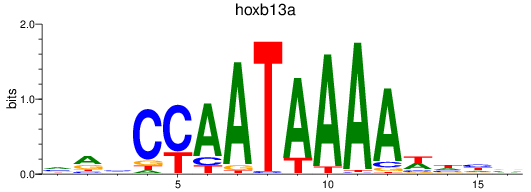
Results for hoxb13a
Z-value: 1.10
Transcription factors associated with hoxb13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb13a
|
ENSDARG00000056015 | homeobox B13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb13a | dr11_v1_chr3_+_23654233_23654233 | 0.94 | 2.0e-04 | Click! |
Activity profile of hoxb13a motif
Sorted Z-values of hoxb13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22675802 | 2.95 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr19_-_5380770 | 2.35 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr7_+_39401388 | 2.26 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr20_-_48485354 | 2.03 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr21_-_25741411 | 2.02 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr10_-_7785930 | 1.84 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr16_-_21785261 | 1.83 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr11_-_18253111 | 1.81 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr22_+_15624371 | 1.71 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr20_+_40150612 | 1.61 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr14_-_24761132 | 1.57 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr6_+_35362225 | 1.50 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr17_-_15382704 | 1.38 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr2_-_59145027 | 1.37 |
ENSDART00000128320
|
FO834803.1
|
|
| chr20_+_29743904 | 1.30 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr5_-_46980651 | 1.29 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr1_-_44940830 | 1.28 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr18_+_44649804 | 1.18 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr7_-_30367650 | 1.17 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr20_-_25709247 | 1.16 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr22_+_11857356 | 1.11 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr8_+_25351863 | 1.10 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr23_-_7799184 | 1.04 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr5_+_22510639 | 1.02 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr17_-_52587598 | 1.00 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr5_+_27434601 | 0.99 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr15_-_28596507 | 0.94 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr4_-_4387012 | 0.85 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr7_+_33130639 | 0.84 |
ENSDART00000142450
ENSDART00000173967 ENSDART00000173832 |
zgc:153219
si:ch211-194p6.7
|
zgc:153219 si:ch211-194p6.7 |
| chr24_-_23323526 | 0.83 |
ENSDART00000112256
ENSDART00000176903 |
zfhx4
|
zinc finger homeobox 4 |
| chr19_-_42557416 | 0.82 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr25_+_25453221 | 0.80 |
ENSDART00000176822
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr16_-_27628994 | 0.73 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr17_-_23412705 | 0.72 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr21_-_37733287 | 0.69 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr1_+_44941031 | 0.69 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr13_-_49802194 | 0.68 |
ENSDART00000148722
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr5_+_34622320 | 0.67 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr1_-_1627487 | 0.66 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr1_-_19079957 | 0.66 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr17_+_21295132 | 0.65 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr12_+_41697664 | 0.65 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr2_-_31735142 | 0.63 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr16_-_563235 | 0.62 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr25_-_35599887 | 0.62 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr8_-_25120231 | 0.60 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr5_-_26181863 | 0.56 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_14390445 | 0.55 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr24_-_23675446 | 0.53 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr19_-_17526735 | 0.53 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr25_+_19106574 | 0.51 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr10_+_39164638 | 0.51 |
ENSDART00000188997
|
CU633908.1
|
|
| chr23_+_30048849 | 0.49 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr25_+_28823952 | 0.48 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr13_+_1182257 | 0.47 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr18_+_1154189 | 0.45 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr9_+_2333550 | 0.45 |
ENSDART00000016417
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr20_-_27733683 | 0.45 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr15_+_45586471 | 0.44 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr23_+_35759843 | 0.44 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr8_+_25352268 | 0.44 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr2_+_45548890 | 0.38 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr12_-_7253270 | 0.37 |
ENSDART00000035762
|
ube2d1b
|
ubiquitin-conjugating enzyme E2D 1b |
| chr3_+_32443395 | 0.35 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr12_-_34758474 | 0.30 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr22_+_18187857 | 0.29 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr24_+_1042594 | 0.29 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr16_+_21738194 | 0.28 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr1_+_8111009 | 0.26 |
ENSDART00000152192
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr1_-_58561963 | 0.23 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr24_-_20808283 | 0.23 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr15_+_11381532 | 0.23 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr7_+_27251376 | 0.22 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr8_+_30709685 | 0.20 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr22_+_18188045 | 0.20 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr2_-_40196547 | 0.19 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr1_+_19515228 | 0.16 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
| chr11_-_29623380 | 0.16 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr5_-_67750907 | 0.15 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr7_-_8324927 | 0.12 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr18_-_9046805 | 0.11 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr4_-_49987980 | 0.11 |
ENSDART00000150428
|
si:dkey-156k2.4
|
si:dkey-156k2.4 |
| chr20_-_52338782 | 0.11 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr1_+_47486104 | 0.09 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr1_-_58562129 | 0.09 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr17_-_45150763 | 0.09 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr1_+_8110562 | 0.09 |
ENSDART00000112160
|
SLC5A10
|
si:dkeyp-9d4.5 |
| chr20_+_34717403 | 0.09 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr20_-_46085840 | 0.08 |
ENSDART00000133714
|
taar12a
|
trace amine associated receptor 12a |
| chr19_-_40776267 | 0.07 |
ENSDART00000189038
|
calcr
|
calcitonin receptor |
| chr4_+_34418211 | 0.04 |
ENSDART00000160070
|
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr17_+_31820401 | 0.04 |
ENSDART00000192607
ENSDART00000157490 |
eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr10_-_22249444 | 0.01 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 2.0 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 1.8 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.5 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.3 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 1.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 2.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.6 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.5 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 2.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |