Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
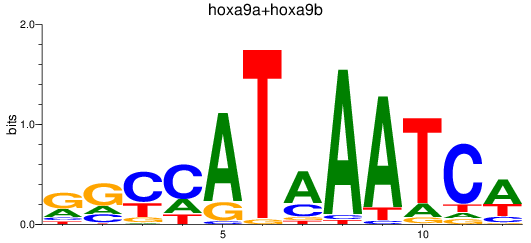
Results for hoxa9a+hoxa9b
Z-value: 0.86
Transcription factors associated with hoxa9a+hoxa9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa9b
|
ENSDARG00000056819 | homeobox A9b |
|
hoxa9a
|
ENSDARG00000105013 | homeobox A9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa9b | dr11_v1_chr16_+_20915319_20915319 | 0.35 | 3.6e-01 | Click! |
| hoxa9a | dr11_v1_chr19_+_19747430_19747430 | 0.23 | 5.5e-01 | Click! |
Activity profile of hoxa9a+hoxa9b motif
Sorted Z-values of hoxa9a+hoxa9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47633438 | 1.38 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr9_-_22299412 | 1.05 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr5_-_32274383 | 0.89 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr3_+_32526263 | 0.81 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr11_-_6048490 | 0.77 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr8_+_6954984 | 0.72 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr5_+_36932718 | 0.71 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr2_+_2223837 | 0.71 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr5_+_32206378 | 0.66 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr7_-_8417315 | 0.62 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr7_-_8374950 | 0.62 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr2_-_5728843 | 0.60 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr1_+_7546259 | 0.59 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr6_+_27151940 | 0.58 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr12_-_26406323 | 0.57 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr3_+_40809011 | 0.56 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr6_-_46875310 | 0.54 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr7_+_39386982 | 0.53 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr15_+_32711172 | 0.50 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr21_-_25685422 | 0.50 |
ENSDART00000182921
|
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr19_-_6385594 | 0.49 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr3_+_33300522 | 0.47 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr22_-_11493236 | 0.45 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr11_-_11266882 | 0.44 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr15_+_33070939 | 0.44 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr12_+_26467847 | 0.43 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr11_-_6188413 | 0.42 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr7_-_4036184 | 0.42 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr2_-_48196092 | 0.41 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr7_+_31879649 | 0.41 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr9_-_296169 | 0.40 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr7_+_48761875 | 0.40 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr12_+_16440708 | 0.39 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr7_-_38659477 | 0.38 |
ENSDART00000138071
|
npsn
|
nephrosin |
| chr10_+_20128267 | 0.38 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr6_+_55032439 | 0.36 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr4_+_21129752 | 0.36 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr10_-_43540196 | 0.36 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr4_-_16354292 | 0.36 |
ENSDART00000139919
|
lum
|
lumican |
| chr7_+_31879986 | 0.35 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_-_31441042 | 0.35 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr10_+_21730585 | 0.34 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr1_-_46981134 | 0.34 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr5_-_51619262 | 0.33 |
ENSDART00000134606
ENSDART00000081249 |
otpb
|
orthopedia homeobox b |
| chr22_+_28446365 | 0.33 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr8_+_26859639 | 0.32 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr10_-_8053753 | 0.32 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr23_-_24263474 | 0.32 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr4_-_27301356 | 0.32 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr4_-_16353733 | 0.31 |
ENSDART00000186785
|
lum
|
lumican |
| chr14_-_24761132 | 0.31 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr16_+_28270037 | 0.31 |
ENSDART00000059035
|
mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr6_+_7444899 | 0.31 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr9_-_6380653 | 0.31 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr22_+_28446557 | 0.30 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr8_+_32402441 | 0.30 |
ENSDART00000191451
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr10_+_37145007 | 0.30 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr9_-_9992697 | 0.30 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr1_-_23157583 | 0.30 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr21_+_39100289 | 0.30 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr16_+_50289916 | 0.30 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr17_+_23300827 | 0.29 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr6_+_41186320 | 0.29 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr21_-_26089964 | 0.29 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr7_-_69853453 | 0.28 |
ENSDART00000049928
|
myoz2a
|
myozenin 2a |
| chr6_-_11768198 | 0.28 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr21_-_43015383 | 0.28 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr13_-_44285793 | 0.28 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr9_+_33009284 | 0.27 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr21_-_27338639 | 0.27 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr20_-_20821783 | 0.27 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr16_+_46111849 | 0.26 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr24_+_38301080 | 0.26 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr9_-_47472998 | 0.25 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr10_-_11385155 | 0.25 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr20_-_29418620 | 0.25 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr5_-_51619742 | 0.25 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr8_+_3405612 | 0.25 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr7_-_38634845 | 0.25 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr8_+_21229718 | 0.25 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr7_-_52950123 | 0.24 |
ENSDART00000009649
|
tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr2_-_3437862 | 0.24 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr19_+_42983613 | 0.24 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr17_-_20717845 | 0.24 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr18_+_808911 | 0.23 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr1_-_18803919 | 0.23 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr7_-_30082931 | 0.23 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr16_+_12836143 | 0.23 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr2_+_16846772 | 0.23 |
ENSDART00000183564
ENSDART00000126718 |
fam131a
|
family with sequence similarity 131, member A |
| chr10_-_35410518 | 0.23 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr4_+_10017049 | 0.23 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr19_-_31042570 | 0.23 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr17_+_51262556 | 0.23 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr11_-_26832685 | 0.23 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr23_-_31506854 | 0.23 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr3_+_34821327 | 0.22 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr12_-_4346085 | 0.22 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr25_-_3830272 | 0.22 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr25_-_7999756 | 0.22 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr3_-_24980067 | 0.22 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr11_-_40147032 | 0.22 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr14_-_31893996 | 0.21 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr5_-_55395384 | 0.21 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr12_+_22404108 | 0.21 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr13_+_13681681 | 0.21 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr12_-_4540564 | 0.21 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr19_+_47290287 | 0.20 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr7_+_69841017 | 0.20 |
ENSDART00000169107
|
FO818704.1
|
|
| chr20_-_32110882 | 0.20 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr16_-_32013913 | 0.20 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr2_+_6926100 | 0.20 |
ENSDART00000153289
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr7_+_48761646 | 0.20 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr17_-_52587598 | 0.20 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr8_-_30424182 | 0.20 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr21_-_25685739 | 0.20 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr2_+_34767171 | 0.20 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr21_-_28920245 | 0.20 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr7_-_24047316 | 0.20 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr7_+_30779761 | 0.19 |
ENSDART00000066806
ENSDART00000173671 |
mcee
|
methylmalonyl CoA epimerase |
| chr8_-_53198154 | 0.19 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr1_+_40023640 | 0.19 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr5_+_26686279 | 0.19 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr14_+_45675306 | 0.19 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr10_+_21650828 | 0.19 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr13_+_19884631 | 0.19 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr1_+_46194333 | 0.19 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr13_-_43599898 | 0.19 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr18_-_42172101 | 0.19 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr13_+_844150 | 0.19 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr7_+_53755054 | 0.19 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr5_+_9382301 | 0.18 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr15_+_8043751 | 0.18 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr10_+_21722892 | 0.18 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr1_+_1805294 | 0.18 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr1_-_681116 | 0.18 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr19_+_43439457 | 0.18 |
ENSDART00000151571
|
ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr16_-_29452039 | 0.18 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr10_-_8053385 | 0.18 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr11_-_141592 | 0.18 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr15_-_18176694 | 0.18 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr5_+_9377005 | 0.18 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr2_-_18830722 | 0.18 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr12_+_18681477 | 0.18 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr11_+_141504 | 0.18 |
ENSDART00000086166
|
NCKAP1L
|
zgc:172352 |
| chr18_+_21122818 | 0.17 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr16_-_8251985 | 0.17 |
ENSDART00000109302
|
fam198a
|
family with sequence similarity 198, member A |
| chr8_-_34065573 | 0.17 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr13_+_25380432 | 0.17 |
ENSDART00000038524
|
gsto1
|
glutathione S-transferase omega 1 |
| chr14_+_35024521 | 0.17 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr23_-_20051369 | 0.17 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr13_+_24834199 | 0.17 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr21_+_11969603 | 0.17 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr10_-_19801821 | 0.17 |
ENSDART00000148013
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr4_-_13255700 | 0.17 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr10_+_26571174 | 0.17 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr18_-_48508585 | 0.17 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr5_-_55395964 | 0.17 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr25_+_32473277 | 0.17 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr11_+_42730639 | 0.16 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr18_-_48517040 | 0.16 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr15_-_452347 | 0.16 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr22_-_31517300 | 0.16 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr16_-_37372613 | 0.16 |
ENSDART00000124090
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr8_-_2616326 | 0.16 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr6_+_21095918 | 0.16 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr5_+_2815021 | 0.16 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr18_+_13792490 | 0.16 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr17_+_15788100 | 0.16 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr23_+_799100 | 0.16 |
ENSDART00000084373
|
frmd4bb
|
FERM domain containing 4Bb |
| chr11_-_25775447 | 0.16 |
ENSDART00000161301
|
plch2b
|
phospholipase C, eta 2b |
| chr9_-_2573121 | 0.16 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr10_-_8046764 | 0.16 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr12_-_30558694 | 0.16 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr1_-_39943596 | 0.16 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr18_+_3004572 | 0.15 |
ENSDART00000190328
|
CABZ01002690.1
|
|
| chr19_+_32321797 | 0.15 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr5_-_71838520 | 0.15 |
ENSDART00000174396
|
CU927890.1
|
|
| chr18_+_11858397 | 0.15 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr12_+_27213733 | 0.15 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr13_-_8692432 | 0.15 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr5_-_29238889 | 0.14 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr16_+_37891735 | 0.14 |
ENSDART00000178753
ENSDART00000142104 |
cep162
|
centrosomal protein 162 |
| chr6_-_51101834 | 0.14 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr17_-_19626357 | 0.14 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr23_+_11669337 | 0.14 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr11_-_44030962 | 0.14 |
ENSDART00000171910
|
FP016005.1
|
|
| chr9_-_2572790 | 0.14 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr15_-_1822166 | 0.14 |
ENSDART00000156883
|
mmp28
|
matrix metallopeptidase 28 |
| chr25_-_13842618 | 0.13 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr11_+_44356707 | 0.13 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr8_-_38159805 | 0.13 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr23_+_40951443 | 0.13 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr4_-_18635005 | 0.13 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr9_+_13714379 | 0.13 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr6_-_29105727 | 0.13 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr1_+_52929185 | 0.13 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr16_-_17175731 | 0.13 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr16_+_33655890 | 0.13 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr2_-_37797577 | 0.13 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr5_-_62940851 | 0.13 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa9a+hoxa9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.6 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.6 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.3 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.7 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.7 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.9 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.2 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.2 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 0.1 | 0.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |