Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
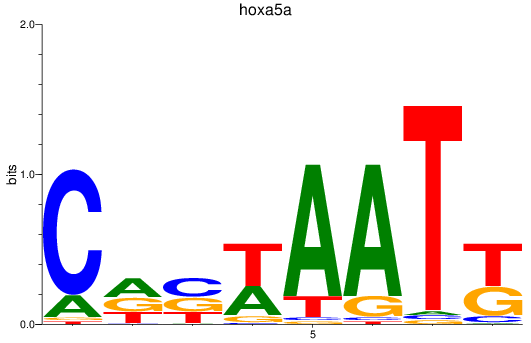
Results for hoxa5a
Z-value: 0.76
Transcription factors associated with hoxa5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa5a
|
ENSDARG00000102501 | homeobox A5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa5a | dr11_v1_chr19_+_19759577_19759577 | -0.82 | 7.2e-03 | Click! |
Activity profile of hoxa5a motif
Sorted Z-values of hoxa5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_55648336 | 1.46 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr22_-_20695237 | 1.19 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr10_-_1961576 | 1.18 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_+_56657804 | 1.09 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr5_-_26247215 | 1.04 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr2_+_37430917 | 1.02 |
ENSDART00000140558
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr17_-_868004 | 0.95 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr23_+_19790962 | 0.95 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr3_+_24595922 | 0.94 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr11_+_18183220 | 0.93 |
ENSDART00000113468
|
LO018315.10
|
|
| chr23_-_3511630 | 0.86 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr3_-_32873641 | 0.84 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr6_-_2134581 | 0.83 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr5_-_26247973 | 0.83 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr17_+_5931530 | 0.82 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr19_-_10920577 | 0.81 |
ENSDART00000165630
|
ecm1a
|
extracellular matrix protein 1a |
| chr8_-_52909850 | 0.77 |
ENSDART00000161943
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr1_+_19602389 | 0.77 |
ENSDART00000088933
ENSDART00000141579 ENSDART00000111555 |
fbxo10
|
F-box protein 10 |
| chr10_-_2942900 | 0.76 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr1_-_44000940 | 0.76 |
ENSDART00000134932
ENSDART00000138447 |
zgc:66472
|
zgc:66472 |
| chr3_+_18807006 | 0.69 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr2_+_5300405 | 0.68 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr1_+_24482406 | 0.64 |
ENSDART00000163140
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr11_-_16021424 | 0.64 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr7_+_71776905 | 0.63 |
ENSDART00000189745
ENSDART00000161344 ENSDART00000158782 |
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr10_+_44700103 | 0.63 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr11_-_11965033 | 0.60 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr16_-_2390931 | 0.59 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr15_-_25613114 | 0.59 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr2_+_54327160 | 0.57 |
ENSDART00000182320
|
LO018508.1
|
|
| chr5_-_16475682 | 0.57 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_+_31791001 | 0.55 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr1_-_55068941 | 0.54 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr23_-_16981899 | 0.54 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr4_-_5775507 | 0.54 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr10_+_44699734 | 0.54 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr11_+_30636351 | 0.54 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr2_+_32846602 | 0.53 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr2_+_37897079 | 0.53 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr20_+_52774730 | 0.53 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr11_-_44999858 | 0.53 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr4_-_20232974 | 0.52 |
ENSDART00000193353
|
stk38l
|
serine/threonine kinase 38 like |
| chr15_-_34871968 | 0.52 |
ENSDART00000191980
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr2_-_31711547 | 0.52 |
ENSDART00000175525
ENSDART00000189763 ENSDART00000113498 |
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr4_+_11723852 | 0.52 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr3_+_16841942 | 0.52 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr25_+_20694177 | 0.51 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr6_+_6662479 | 0.51 |
ENSDART00000065568
|
zgc:113227
|
zgc:113227 |
| chr13_-_28688104 | 0.50 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr2_-_21819421 | 0.49 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr11_+_43751263 | 0.49 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr14_-_26498196 | 0.49 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr7_-_40630698 | 0.49 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr6_-_41135215 | 0.49 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr23_+_32028574 | 0.49 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr9_-_34937025 | 0.49 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr7_+_38809241 | 0.49 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr15_+_26940569 | 0.49 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr3_+_17346502 | 0.48 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr3_+_27606505 | 0.48 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr24_-_31123365 | 0.48 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr14_+_10981703 | 0.47 |
ENSDART00000091169
|
abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr24_+_39277043 | 0.47 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr16_+_40954481 | 0.46 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr5_+_31283576 | 0.45 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr2_+_8779164 | 0.44 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr24_+_5789790 | 0.43 |
ENSDART00000189600
|
BX470065.1
|
|
| chr19_+_11432330 | 0.42 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr8_-_22274222 | 0.42 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr19_-_47832853 | 0.42 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr9_-_12269847 | 0.41 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr9_-_746317 | 0.41 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr21_-_44772738 | 0.40 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr4_-_14162327 | 0.40 |
ENSDART00000138656
|
si:dkey-234l24.1
|
si:dkey-234l24.1 |
| chr24_+_17349177 | 0.39 |
ENSDART00000176338
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr16_+_23531583 | 0.39 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr19_+_19241372 | 0.38 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr5_+_71924175 | 0.37 |
ENSDART00000115182
ENSDART00000170215 |
nup214
|
nucleoporin 214 |
| chr5_+_8964926 | 0.37 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr25_-_10622449 | 0.37 |
ENSDART00000155375
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr9_-_30220818 | 0.36 |
ENSDART00000140929
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr11_-_38083397 | 0.35 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr21_-_35325466 | 0.34 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr17_-_10738001 | 0.33 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr5_+_27488975 | 0.33 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr10_-_13343831 | 0.33 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr1_+_6189949 | 0.32 |
ENSDART00000092277
|
retreg2
|
reticulophagy regulator family member 2 |
| chr8_-_30779101 | 0.32 |
ENSDART00000062229
|
p2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr22_-_9896180 | 0.31 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr4_-_14173138 | 0.31 |
ENSDART00000142487
|
si:dkey-234l24.7
|
si:dkey-234l24.7 |
| chr2_-_38204845 | 0.30 |
ENSDART00000142342
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr15_-_23692359 | 0.29 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr3_-_36750068 | 0.29 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr21_-_40413191 | 0.28 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr22_+_5118361 | 0.28 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr15_-_15230264 | 0.27 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr14_-_30540967 | 0.27 |
ENSDART00000026907
|
her11
|
hairy-related 11 |
| chr12_-_31461412 | 0.27 |
ENSDART00000175929
ENSDART00000186342 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr2_+_11031360 | 0.27 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr5_+_66433287 | 0.27 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr19_+_19600297 | 0.26 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr22_-_11517377 | 0.26 |
ENSDART00000193885
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr8_+_47571211 | 0.26 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr18_+_13300768 | 0.26 |
ENSDART00000137506
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr17_-_8656155 | 0.25 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr2_-_33645411 | 0.24 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr2_-_32826108 | 0.24 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr21_-_2348838 | 0.24 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr5_+_4436405 | 0.23 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr24_+_31374324 | 0.23 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr21_+_22400951 | 0.22 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr12_+_10443785 | 0.21 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr6_-_9236309 | 0.20 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr11_+_10975481 | 0.20 |
ENSDART00000160488
|
itgb6
|
integrin, beta 6 |
| chr2_+_32826235 | 0.20 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr3_-_59981476 | 0.19 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr15_+_8335600 | 0.19 |
ENSDART00000190066
ENSDART00000152603 |
egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr11_-_3613558 | 0.19 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr3_+_59880317 | 0.19 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr22_+_1462177 | 0.19 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr21_-_32467799 | 0.18 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr21_+_11885404 | 0.18 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr3_+_32112004 | 0.18 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr15_-_47956388 | 0.18 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr5_-_23909934 | 0.17 |
ENSDART00000142516
|
si:ch211-135f11.1
|
si:ch211-135f11.1 |
| chr23_-_18381361 | 0.17 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr14_-_33859149 | 0.16 |
ENSDART00000163877
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr5_-_67783593 | 0.15 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr8_+_35212233 | 0.15 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr4_-_11403811 | 0.15 |
ENSDART00000067272
ENSDART00000140018 |
pimr173
|
Pim proto-oncogene, serine/threonine kinase, related 173 |
| chr14_-_16151055 | 0.14 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr25_-_3627130 | 0.14 |
ENSDART00000171863
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr15_-_1844048 | 0.12 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_+_47290287 | 0.12 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr15_-_16679517 | 0.12 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr4_-_65037243 | 0.12 |
ENSDART00000170059
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr23_+_4324625 | 0.12 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr21_-_2952789 | 0.11 |
ENSDART00000182898
|
znf971
|
zinc finger protein 971 |
| chr19_-_47570672 | 0.11 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr10_+_5025696 | 0.11 |
ENSDART00000031165
|
enoph1
|
enolase-phosphatase 1 |
| chr3_-_32898626 | 0.10 |
ENSDART00000103201
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr14_-_30490763 | 0.09 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr9_+_21843915 | 0.09 |
ENSDART00000101977
|
kctd12.1
|
potassium channel tetramerisation domain containing 12.1 |
| chr23_+_40109353 | 0.09 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr5_+_59392183 | 0.09 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr8_-_11146924 | 0.08 |
ENSDART00000126824
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr9_+_44721808 | 0.08 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr22_+_2844865 | 0.08 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr17_+_30751462 | 0.07 |
ENSDART00000154184
|
ldah
|
lipid droplet associated hydrolase |
| chr8_-_36287046 | 0.07 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr17_-_200316 | 0.06 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr15_+_14141045 | 0.06 |
ENSDART00000154635
|
si:ch211-116o3.3
|
si:ch211-116o3.3 |
| chr21_+_28749720 | 0.06 |
ENSDART00000145178
|
zgc:100829
|
zgc:100829 |
| chr23_-_6765653 | 0.06 |
ENSDART00000192310
|
FP102169.1
|
|
| chr16_-_25085327 | 0.06 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr11_-_14813029 | 0.06 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr13_-_37383625 | 0.06 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr1_-_9940494 | 0.06 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr17_-_43391499 | 0.05 |
ENSDART00000189280
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr2_-_37803614 | 0.05 |
ENSDART00000154124
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr21_-_2299002 | 0.05 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr16_-_48074540 | 0.05 |
ENSDART00000187258
ENSDART00000186694 |
CSMD3 (1 of many)
|
si:ch211-236p22.1 |
| chr11_-_7156620 | 0.05 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr5_+_63857055 | 0.05 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr2_-_43942595 | 0.05 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr12_+_8003872 | 0.05 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr4_+_28997595 | 0.04 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr10_+_35468978 | 0.04 |
ENSDART00000131984
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr16_+_25664387 | 0.04 |
ENSDART00000184394
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr1_-_45614481 | 0.04 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_31254979 | 0.03 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr21_+_5580948 | 0.03 |
ENSDART00000160373
|
ly6m7
|
lymphocyte antigen 6 family member M7 |
| chr16_-_12787029 | 0.03 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr3_+_32365811 | 0.03 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr21_+_35327589 | 0.02 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr18_+_41313999 | 0.02 |
ENSDART00000048985
|
veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr23_-_44470253 | 0.02 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr6_-_39489190 | 0.02 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr3_-_3723494 | 0.01 |
ENSDART00000056023
|
si:dkey-217f16.1
|
si:dkey-217f16.1 |
| chr8_-_50888806 | 0.01 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr22_-_18387059 | 0.01 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr16_+_25664078 | 0.01 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr7_-_6604623 | 0.01 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr8_-_11146687 | 0.01 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr16_-_35394422 | 0.00 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr4_-_72101234 | 0.00 |
ENSDART00000167048
|
slco1f1
|
solute carrier organic anion transporter family, member 1F1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.4 | 1.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.6 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 1.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:2000191 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.5 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.5 | GO:0048796 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.3 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.4 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 1.1 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.2 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |