Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
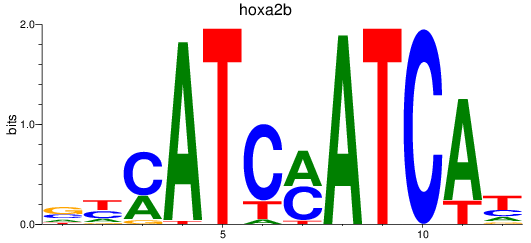
Results for hoxa2b
Z-value: 0.61
Transcription factors associated with hoxa2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa2b
|
ENSDARG00000023031 | homeobox A2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa2b | dr11_v1_chr16_+_20926673_20926673 | 0.44 | 2.4e-01 | Click! |
Activity profile of hoxa2b motif
Sorted Z-values of hoxa2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 1.87 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr9_-_22135420 | 0.70 |
ENSDART00000184959
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr9_-_45601103 | 0.63 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr25_+_20215964 | 0.62 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr9_-_22099536 | 0.62 |
ENSDART00000101923
|
CR391987.1
|
|
| chr25_+_20216159 | 0.56 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr7_+_29952719 | 0.53 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr12_-_11457625 | 0.52 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr3_-_32818607 | 0.52 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr7_-_8374950 | 0.52 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr9_-_22232902 | 0.50 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr9_-_22135576 | 0.49 |
ENSDART00000101902
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr14_-_36378494 | 0.42 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr23_+_5490854 | 0.39 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr1_+_17676745 | 0.38 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr7_-_8417315 | 0.38 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr9_-_44295071 | 0.37 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr9_-_22310919 | 0.36 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr5_+_37837245 | 0.36 |
ENSDART00000171617
|
epd
|
ependymin |
| chr12_-_25916530 | 0.35 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr2_-_722156 | 0.34 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr4_+_12931763 | 0.33 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr4_-_16333944 | 0.33 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr23_-_18609475 | 0.32 |
ENSDART00000104524
|
zgc:158296
|
zgc:158296 |
| chr19_+_1835940 | 0.32 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr14_+_25465346 | 0.31 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr22_-_600016 | 0.31 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr4_+_21129752 | 0.31 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr2_+_47623202 | 0.30 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr1_+_16144615 | 0.30 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr7_-_24472991 | 0.30 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr5_+_32221755 | 0.30 |
ENSDART00000125917
|
myhc4
|
myosin heavy chain 4 |
| chr13_+_1726953 | 0.29 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr5_+_36932718 | 0.29 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr12_-_26406323 | 0.29 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr25_+_19105804 | 0.28 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr2_-_2020044 | 0.27 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr3_+_23752150 | 0.27 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr21_-_43079161 | 0.27 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr16_+_20904754 | 0.27 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr16_+_12022543 | 0.27 |
ENSDART00000012673
|
gnb3a
|
guanine nucleotide binding protein (G protein), beta polypeptide 3a |
| chr16_-_45058919 | 0.27 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr13_+_23282549 | 0.27 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr10_-_39011514 | 0.27 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr23_-_11870962 | 0.26 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr12_+_48241841 | 0.26 |
ENSDART00000168616
|
ppa1a
|
pyrophosphatase (inorganic) 1a |
| chr16_-_45225520 | 0.26 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr11_-_22605981 | 0.26 |
ENSDART00000186923
|
myog
|
myogenin |
| chr1_-_46981134 | 0.26 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr6_+_29791164 | 0.26 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr8_-_16697912 | 0.25 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr18_+_13792490 | 0.25 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr25_-_13363286 | 0.25 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr1_-_26294995 | 0.24 |
ENSDART00000168594
|
cxxc4
|
CXXC finger 4 |
| chr12_+_39685485 | 0.24 |
ENSDART00000163403
|
LO017650.1
|
|
| chr2_+_27010439 | 0.24 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr4_+_76575585 | 0.24 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr8_+_22931427 | 0.24 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr5_-_20194876 | 0.24 |
ENSDART00000122587
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr24_-_6897884 | 0.24 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr1_-_59252973 | 0.24 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr22_+_12770877 | 0.24 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr6_+_15268685 | 0.23 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr23_+_4689626 | 0.23 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr10_+_20128267 | 0.23 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr7_+_26629084 | 0.23 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr10_-_20453995 | 0.23 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr14_+_34486629 | 0.23 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr10_-_20445549 | 0.22 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr4_+_17279966 | 0.22 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr21_+_6751405 | 0.22 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr3_+_40809011 | 0.22 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr20_-_8443425 | 0.22 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_-_12052132 | 0.22 |
ENSDART00000074361
|
zgc:110789
|
zgc:110789 |
| chr1_+_46194333 | 0.22 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr2_-_31634978 | 0.21 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr7_-_13882988 | 0.21 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr20_+_23238833 | 0.21 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr15_+_32268790 | 0.21 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr14_-_52542928 | 0.21 |
ENSDART00000075749
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr14_+_13454840 | 0.20 |
ENSDART00000161854
|
pls3
|
plastin 3 (T isoform) |
| chr12_-_4346085 | 0.20 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr23_+_10146542 | 0.20 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr20_-_9462433 | 0.20 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr21_-_28920245 | 0.20 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr11_-_7320211 | 0.20 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr16_+_10776688 | 0.20 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr10_+_439692 | 0.19 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr1_-_10071422 | 0.19 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr13_+_29770837 | 0.19 |
ENSDART00000076998
|
pax2a
|
paired box 2a |
| chr7_+_46252993 | 0.19 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr24_+_119680 | 0.19 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr17_-_20897250 | 0.19 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr18_-_42785469 | 0.19 |
ENSDART00000024768
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr24_-_26310854 | 0.19 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr10_-_26744131 | 0.19 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr21_-_44512893 | 0.19 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr13_+_9432501 | 0.19 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr25_-_31396479 | 0.18 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr5_-_20195350 | 0.18 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr11_+_26476153 | 0.18 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr9_+_48007081 | 0.18 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr1_+_52929185 | 0.18 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr18_+_15706160 | 0.17 |
ENSDART00000131524
|
si:ch211-264e16.1
|
si:ch211-264e16.1 |
| chr3_+_27027781 | 0.17 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr4_+_26496489 | 0.17 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr17_-_49800869 | 0.17 |
ENSDART00000156264
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr13_+_22249636 | 0.17 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr9_+_22003942 | 0.17 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr1_+_16127825 | 0.17 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr8_+_28527776 | 0.17 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr3_-_26109322 | 0.17 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr16_+_46111849 | 0.17 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr7_+_20503344 | 0.17 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr10_+_35417099 | 0.17 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr4_-_5597802 | 0.17 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr12_+_5189776 | 0.17 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr13_-_5569562 | 0.16 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr4_-_16412084 | 0.16 |
ENSDART00000188460
|
dcn
|
decorin |
| chr7_+_35229645 | 0.16 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr24_-_6898302 | 0.16 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr19_-_48312109 | 0.16 |
ENSDART00000161103
|
si:ch73-359m17.9
|
si:ch73-359m17.9 |
| chr1_-_51465730 | 0.16 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr18_-_46010 | 0.16 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr20_-_53992952 | 0.16 |
ENSDART00000170138
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr11_-_21586157 | 0.16 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr22_+_34701848 | 0.16 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr20_-_35841570 | 0.16 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr15_-_26552652 | 0.16 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr13_-_31435137 | 0.15 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr10_+_21511495 | 0.15 |
ENSDART00000178395
|
BX957322.3
|
|
| chr15_+_45640906 | 0.15 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr10_+_34377697 | 0.15 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr25_+_15354095 | 0.15 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr19_-_42607451 | 0.15 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr8_-_54223316 | 0.15 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr16_-_54942532 | 0.15 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr23_+_36095260 | 0.15 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr9_+_17438765 | 0.15 |
ENSDART00000138953
|
rgcc
|
regulator of cell cycle |
| chr21_-_27338639 | 0.15 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr8_-_20380750 | 0.15 |
ENSDART00000113910
|
adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr18_-_8877077 | 0.15 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr24_+_13735616 | 0.15 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr22_-_33679277 | 0.14 |
ENSDART00000169948
|
FO904977.1
|
|
| chr25_-_18953322 | 0.14 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr25_+_21833287 | 0.14 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr3_+_33300522 | 0.14 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr13_-_1349922 | 0.14 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr23_-_19500559 | 0.14 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr10_+_3875716 | 0.14 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr11_-_1509773 | 0.14 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr21_+_6961635 | 0.14 |
ENSDART00000137281
|
si:ch211-93g21.2
|
si:ch211-93g21.2 |
| chr7_-_51321126 | 0.14 |
ENSDART00000067647
|
rasl11a
|
RAS-like, family 11, member A |
| chr1_-_25966068 | 0.14 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr3_+_23726148 | 0.14 |
ENSDART00000174580
|
hoxb3a
|
homeobox B3a |
| chr7_-_34475696 | 0.14 |
ENSDART00000058095
|
si:zfos-1897c11.1
|
si:zfos-1897c11.1 |
| chr2_-_26001173 | 0.14 |
ENSDART00000010615
|
cldn11a
|
claudin 11a |
| chr5_+_36693859 | 0.14 |
ENSDART00000019259
|
dlb
|
deltaB |
| chr6_-_29105727 | 0.14 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr16_-_37964325 | 0.14 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr14_+_109016 | 0.14 |
ENSDART00000158405
|
gpc2
|
glypican 2 |
| chr23_+_6795709 | 0.14 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr13_-_21688176 | 0.14 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr4_-_2545310 | 0.14 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr11_+_25472758 | 0.14 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr16_-_31188715 | 0.14 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr18_-_48745517 | 0.14 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr16_-_54455573 | 0.14 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr4_-_25485404 | 0.13 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr12_-_28881638 | 0.13 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr7_-_20453661 | 0.13 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr10_+_7798087 | 0.13 |
ENSDART00000141824
|
sftpba
|
surfactant protein Ba |
| chr1_+_29281764 | 0.13 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr13_-_32577386 | 0.13 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr20_+_19512727 | 0.13 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr18_+_30847237 | 0.13 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr3_+_60721342 | 0.13 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr23_+_2361184 | 0.13 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr23_-_20309505 | 0.13 |
ENSDART00000130856
|
lamb2l
|
laminin, beta 2-like |
| chr8_+_44420108 | 0.13 |
ENSDART00000075381
|
CU571323.1
|
|
| chr8_-_42624454 | 0.13 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr12_+_48634927 | 0.13 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr22_+_20208185 | 0.13 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr7_+_73593814 | 0.13 |
ENSDART00000110544
|
znf219
|
zinc finger protein 219 |
| chr17_-_49978986 | 0.13 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr6_-_16406210 | 0.13 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr11_+_36243774 | 0.13 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr21_-_40174647 | 0.13 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr10_+_17850934 | 0.13 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr10_-_4316015 | 0.13 |
ENSDART00000109916
ENSDART00000180867 |
grin3a
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
| chr20_+_7243879 | 0.13 |
ENSDART00000122298
|
bsnd
|
barttin CLCNK-type chloride channel accessory beta subunit |
| chr16_-_35952789 | 0.12 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr17_-_10838434 | 0.12 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr5_+_64842730 | 0.12 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr15_+_45643787 | 0.12 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr6_+_41186320 | 0.12 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr23_-_44226556 | 0.12 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr22_+_24715282 | 0.12 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr24_-_23942722 | 0.12 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr16_+_1254390 | 0.12 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.1 | 0.4 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.4 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.2 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.3 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.2 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.4 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.0 | GO:0071635 | regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.2 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.4 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.1 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.0 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 2.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.3 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.3 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.1 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 1.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.1 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |