Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
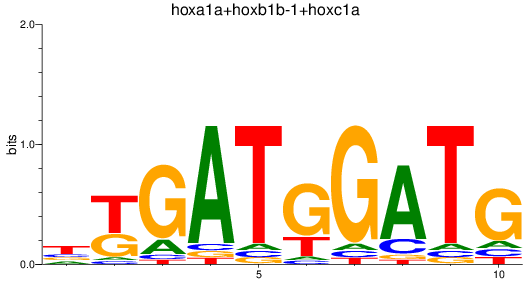
Results for hoxa1a+hoxb1b-1+hoxc1a
Z-value: 1.47
Transcription factors associated with hoxa1a+hoxb1b-1+hoxc1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb1b-1
|
ENSDARG00000054033 | homeobox B1b |
|
hoxc1a
|
ENSDARG00000070337 | homeobox C1a |
|
hoxa1a
|
ENSDARG00000104307 | homeobox A1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc1a | dr11_v1_chr23_+_36178104_36178104 | 0.89 | 1.2e-03 | Click! |
| hoxa1a | dr11_v1_chr19_+_19786117_19786213 | 0.88 | 2.0e-03 | Click! |
| hoxb1b | dr11_v1_chr12_+_27141140_27141140 | 0.75 | 2.0e-02 | Click! |
Activity profile of hoxa1a+hoxb1b-1+hoxc1a motif
Sorted Z-values of hoxa1a+hoxb1b-1+hoxc1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_1383914 | 4.88 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr3_-_55147731 | 4.66 |
ENSDART00000155871
ENSDART00000109016 ENSDART00000122904 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr12_-_25916530 | 4.31 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr7_+_29952719 | 3.28 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr25_+_20215964 | 2.77 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr6_+_29791164 | 2.67 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr21_-_26495700 | 2.60 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr20_-_32446406 | 2.52 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr19_-_45985989 | 2.49 |
ENSDART00000165897
|
si:ch211-153f2.3
|
si:ch211-153f2.3 |
| chr25_+_20216159 | 2.43 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr13_-_39160018 | 2.39 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr19_-_2231146 | 2.29 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr18_-_23874929 | 2.09 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_+_40684 | 2.03 |
ENSDART00000164231
|
CABZ01111555.1
|
|
| chr8_-_40464935 | 2.01 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr18_-_23875370 | 1.89 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr20_-_9462433 | 1.88 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr18_+_24919614 | 1.74 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr22_-_34937455 | 1.66 |
ENSDART00000169217
ENSDART00000188330 ENSDART00000165142 |
slit1b
|
slit homolog 1b (Drosophila) |
| chr4_+_4816695 | 1.63 |
ENSDART00000136962
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr22_-_600016 | 1.56 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr11_-_1131569 | 1.50 |
ENSDART00000173228
|
slc6a11b
|
solute carrier family 6 (neurotransmitter transporter), member 11b |
| chr18_-_23875219 | 1.43 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_-_66028714 | 1.42 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr16_-_31188715 | 1.41 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr19_+_42219165 | 1.41 |
ENSDART00000163192
|
CU896644.1
|
|
| chr23_-_12015139 | 1.37 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr22_-_15602593 | 1.36 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr3_+_23752150 | 1.36 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr14_+_34547554 | 1.30 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr13_+_35745572 | 1.29 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr11_-_1509773 | 1.27 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr20_-_35841570 | 1.26 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr23_-_11870962 | 1.23 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr21_-_43079161 | 1.23 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr22_-_15602760 | 1.21 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr10_-_22127942 | 1.20 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr13_-_43149063 | 1.19 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr20_-_29483514 | 1.19 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr16_+_46111849 | 1.17 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr9_+_34425736 | 1.16 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr25_+_7461624 | 1.14 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr5_-_55600689 | 1.13 |
ENSDART00000013229
|
gnaq
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr6_+_52064304 | 1.10 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr23_-_12014931 | 1.07 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr9_-_23217196 | 1.06 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr21_-_31290582 | 1.05 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr14_-_7888748 | 1.05 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr14_+_24215046 | 1.03 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr19_-_47555956 | 1.01 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr18_+_24922125 | 1.01 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr11_-_11471857 | 1.00 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr23_+_36178104 | 1.00 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr18_-_42785469 | 1.00 |
ENSDART00000024768
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr16_-_13730152 | 0.98 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr20_+_26349002 | 0.98 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_+_23768898 | 0.96 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr24_+_119680 | 0.96 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr4_+_25706037 | 0.89 |
ENSDART00000141133
|
lamb1b
|
laminin, beta 1b |
| chr20_+_27691307 | 0.87 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr14_+_2478994 | 0.84 |
ENSDART00000170538
|
CU929317.1
|
|
| chr4_+_11384891 | 0.84 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr11_-_21528056 | 0.84 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr20_-_47550577 | 0.80 |
ENSDART00000187361
|
CABZ01053323.1
|
|
| chr8_-_10932206 | 0.80 |
ENSDART00000124313
|
nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr12_+_25945560 | 0.78 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr6_-_13401133 | 0.77 |
ENSDART00000151206
|
fmnl2b
|
formin-like 2b |
| chr8_-_7440364 | 0.76 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr10_+_439692 | 0.76 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr4_+_21129752 | 0.76 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr20_-_25369767 | 0.73 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr13_-_32577386 | 0.72 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr25_-_24074500 | 0.71 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr2_-_31754292 | 0.71 |
ENSDART00000192498
|
ralyl
|
RALY RNA binding protein like |
| chr7_+_25036188 | 0.70 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr3_+_15805917 | 0.70 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr6_+_23752593 | 0.69 |
ENSDART00000164366
|
zgc:158654
|
zgc:158654 |
| chr5_-_52921532 | 0.66 |
ENSDART00000160566
|
CU695117.1
|
|
| chr9_+_11202552 | 0.66 |
ENSDART00000151931
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr17_+_51764310 | 0.63 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr8_+_18830759 | 0.62 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr2_-_45630823 | 0.61 |
ENSDART00000183553
|
CR450720.1
|
|
| chr15_+_59417 | 0.60 |
ENSDART00000187260
|
AXL
|
AXL receptor tyrosine kinase |
| chr5_-_22544151 | 0.60 |
ENSDART00000006474
|
glra4b
|
glycine receptor, alpha 4b |
| chr7_+_30371893 | 0.60 |
ENSDART00000075513
|
aqp9b
|
aquaporin 9b |
| chr10_-_31175744 | 0.60 |
ENSDART00000191728
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr6_-_29105727 | 0.59 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr17_+_22956427 | 0.57 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr24_+_34113424 | 0.56 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr3_-_49925313 | 0.54 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr16_+_46401576 | 0.54 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr4_-_16641742 | 0.54 |
ENSDART00000180605
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr9_-_51370293 | 0.54 |
ENSDART00000084806
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr10_-_27741793 | 0.53 |
ENSDART00000129369
ENSDART00000192440 ENSDART00000189808 ENSDART00000138149 |
auts2a
si:dkey-33o22.1
|
autism susceptibility candidate 2a si:dkey-33o22.1 |
| chr25_-_36827491 | 0.53 |
ENSDART00000170941
|
CU929365.1
|
|
| chr21_+_43561650 | 0.52 |
ENSDART00000085071
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr7_-_72067475 | 0.49 |
ENSDART00000017763
|
LO018380.1
|
|
| chr16_+_14707960 | 0.48 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr4_-_14926637 | 0.47 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr14_+_19258702 | 0.46 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr8_+_6410933 | 0.46 |
ENSDART00000168789
|
lrrtm4l2
|
leucine rich repeat transmembrane neuronal 4 like 2 |
| chr7_+_16835218 | 0.45 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr15_+_15456536 | 0.44 |
ENSDART00000109004
ENSDART00000156835 |
zgc:174895
|
zgc:174895 |
| chr25_-_29987839 | 0.44 |
ENSDART00000154088
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr22_+_635813 | 0.43 |
ENSDART00000179067
|
CU856139.1
|
|
| chr17_-_6202816 | 0.41 |
ENSDART00000151908
|
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr11_-_11458208 | 0.41 |
ENSDART00000005485
|
krt93
|
keratin 93 |
| chr13_-_41908583 | 0.41 |
ENSDART00000136515
|
ipmka
|
inositol polyphosphate multikinase a |
| chr5_-_52277643 | 0.40 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr12_+_39685485 | 0.36 |
ENSDART00000163403
|
LO017650.1
|
|
| chr17_-_45009782 | 0.36 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr4_-_13626831 | 0.35 |
ENSDART00000173077
|
si:dkeyp-81f3.4
|
si:dkeyp-81f3.4 |
| chr1_+_10221568 | 0.35 |
ENSDART00000152424
|
npy2rl
|
neuropeptide Y receptor Y2, like |
| chr6_-_55585423 | 0.32 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr22_+_3232925 | 0.31 |
ENSDART00000166754
|
CU929402.1
|
|
| chr13_-_51065048 | 0.31 |
ENSDART00000168085
|
CABZ01078663.1
|
|
| chr6_-_16561536 | 0.30 |
ENSDART00000167295
|
unc80
|
unc-80 homolog (C. elegans) |
| chr18_-_48755037 | 0.29 |
ENSDART00000109673
|
tirap
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr23_+_21067684 | 0.29 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr24_+_34806822 | 0.29 |
ENSDART00000148407
ENSDART00000188328 |
mchr2
|
melanin-concentrating hormone receptor 2 |
| chr17_+_8183393 | 0.29 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr17_+_20607487 | 0.28 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr9_-_48296469 | 0.28 |
ENSDART00000058255
|
bbs5
|
Bardet-Biedl syndrome 5 |
| chr15_+_15456910 | 0.27 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr13_-_2189761 | 0.26 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr1_-_47431453 | 0.26 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr17_-_26604946 | 0.26 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr10_-_26766780 | 0.26 |
ENSDART00000146666
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr4_-_14926106 | 0.25 |
ENSDART00000147629
|
prdm4
|
PR domain containing 4 |
| chr18_+_41542542 | 0.23 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr7_+_31074636 | 0.23 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr3_-_214709 | 0.23 |
ENSDART00000098311
|
CABZ01079818.1
|
|
| chr17_-_20394566 | 0.22 |
ENSDART00000154667
|
sorcs3b
|
sortilin related VPS10 domain containing receptor 3b |
| chr13_-_28272299 | 0.21 |
ENSDART00000006393
|
tlx1
|
T cell leukemia homeobox 1 |
| chr1_+_20069535 | 0.20 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr17_-_19626357 | 0.19 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr16_-_17300030 | 0.18 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr8_-_44015210 | 0.17 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr15_-_27972474 | 0.16 |
ENSDART00000162753
|
CR391930.1
|
|
| chr16_+_25196572 | 0.16 |
ENSDART00000141956
|
tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr2_-_16217344 | 0.15 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_21787507 | 0.14 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr2_+_6303639 | 0.13 |
ENSDART00000132859
|
otol1a
|
otolin 1a |
| chr23_-_333457 | 0.13 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr4_-_71983381 | 0.13 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr18_-_16392448 | 0.13 |
ENSDART00000180253
|
mgat4c
|
mgat4 family, member C |
| chr17_+_45009868 | 0.12 |
ENSDART00000085009
|
coq6
|
coenzyme Q6 monooxygenase |
| chr11_+_33628104 | 0.11 |
ENSDART00000165318
|
thsd7bb
|
thrombospondin, type I, domain containing 7Bb |
| chr9_+_22351443 | 0.10 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr13_+_37273010 | 0.10 |
ENSDART00000144387
|
gphb5
|
glycoprotein hormone beta 5 |
| chr25_+_12849609 | 0.10 |
ENSDART00000168144
|
ccl33.2
|
chemokine (C-C motif) ligand 33, duplicate 2 |
| chr3_+_46628885 | 0.10 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr17_-_26604549 | 0.09 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr22_+_38778649 | 0.09 |
ENSDART00000075873
|
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr8_-_52745141 | 0.09 |
ENSDART00000168359
ENSDART00000168252 |
fgf17
|
fibroblast growth factor 17 |
| chr20_-_28842524 | 0.09 |
ENSDART00000046035
ENSDART00000139843 ENSDART00000129858 ENSDART00000137425 ENSDART00000135720 |
max
|
myc associated factor X |
| chr16_-_20932896 | 0.09 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr5_+_61976511 | 0.07 |
ENSDART00000050885
|
ehd2a
|
EH-domain containing 2a |
| chr7_-_7810348 | 0.07 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr5_+_61301525 | 0.06 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr24_+_26039464 | 0.06 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr4_+_74226447 | 0.06 |
ENSDART00000174336
ENSDART00000191455 ENSDART00000180062 |
ptprr
|
protein tyrosine phosphatase, receptor type, r |
| chr7_-_18617187 | 0.05 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr22_+_38778854 | 0.04 |
ENSDART00000182926
ENSDART00000125466 |
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr8_-_39739056 | 0.02 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr16_-_16761164 | 0.02 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr12_-_36268723 | 0.02 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_49481672 | 0.01 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr2_+_22765492 | 0.00 |
ENSDART00000133222
|
espnlb
|
espin like b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa1a+hoxb1b-1+hoxc1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.5 | 2.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 1.3 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.4 | 2.2 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.3 | 2.0 | GO:0055014 | ventricular cardiac myofibril assembly(GO:0055005) atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 1.0 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.2 | 0.7 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.2 | 4.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 4.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 0.8 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 1.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 5.2 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 0.6 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 4.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 2.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 3.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.2 | GO:0071635 | negative regulation of B cell proliferation(GO:0030889) regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) negative regulation of B cell activation(GO:0050869) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 1.7 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 1.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 3.2 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 1.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.1 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0097324 | adult behavior(GO:0030534) melanocyte migration(GO:0097324) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.0 | 0.5 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.0 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 3.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 4.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 0.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 5.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0045335 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.9 | 2.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 5.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 5.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 4.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 2.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 4.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.2 | 0.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 1.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.6 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 1.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.5 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 2.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 2.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 5.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 5.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |