Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
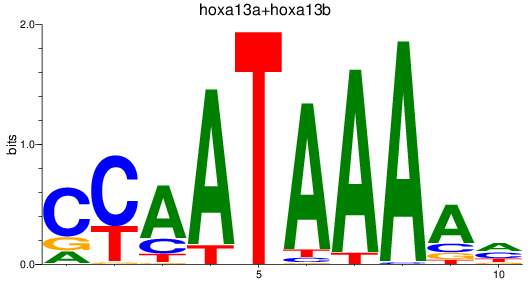
Results for hoxa13a+hoxa13b
Z-value: 1.34
Transcription factors associated with hoxa13a+hoxa13b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa13b
|
ENSDARG00000036254 | homeobox A13b |
|
hoxa13a
|
ENSDARG00000100312 | homeobox A13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa13a | dr11_v1_chr19_+_19729506_19729506 | -0.23 | 5.5e-01 | Click! |
| hoxa13b | dr11_v1_chr16_+_20895904_20895904 | -0.22 | 5.6e-01 | Click! |
Activity profile of hoxa13a+hoxa13b motif
Sorted Z-values of hoxa13a+hoxa13b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_14191592 | 2.08 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr13_+_22675802 | 1.67 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr16_-_45917322 | 1.54 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr23_+_39606108 | 1.32 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr7_-_30367650 | 1.21 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr13_-_50614639 | 1.20 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr19_-_24555623 | 1.19 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr22_+_25590391 | 1.00 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr14_-_21123551 | 0.89 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr7_-_69025306 | 0.86 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr19_-_24555935 | 0.86 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr10_-_24391030 | 0.84 |
ENSDART00000180013
ENSDART00000177914 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr25_+_34641536 | 0.84 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr5_+_68807170 | 0.84 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr19_-_1948236 | 0.81 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr21_-_37733287 | 0.81 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr14_+_34495216 | 0.77 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_+_35362225 | 0.77 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr10_-_6427362 | 0.76 |
ENSDART00000166774
|
ca9
|
carbonic anhydrase IX |
| chr18_-_14677936 | 0.76 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr19_-_42291355 | 0.75 |
ENSDART00000150919
ENSDART00000151034 |
si:ch211-191i18.2
|
si:ch211-191i18.2 |
| chr17_+_27456804 | 0.75 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr23_+_32499916 | 0.75 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr3_+_57948377 | 0.75 |
ENSDART00000047418
|
notum1a
|
notum, palmitoleoyl-protein carboxylesterase a |
| chr3_-_37759969 | 0.72 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr24_+_9475809 | 0.72 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr23_+_7379728 | 0.72 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr4_-_61651223 | 0.69 |
ENSDART00000172688
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr18_+_26422124 | 0.67 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr13_+_30804367 | 0.67 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr24_-_8732519 | 0.65 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr5_-_41307550 | 0.65 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr8_-_19280856 | 0.63 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr25_-_31739309 | 0.63 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr25_+_24616717 | 0.62 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr10_-_6454386 | 0.61 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr7_+_67429185 | 0.61 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr7_-_16562200 | 0.61 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr13_+_28495983 | 0.60 |
ENSDART00000144968
|
fgf8a
|
fibroblast growth factor 8a |
| chr9_-_53666031 | 0.60 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr21_-_34844316 | 0.60 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr3_-_48259289 | 0.59 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr5_-_36328688 | 0.59 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr15_+_14856307 | 0.59 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr14_+_34490445 | 0.58 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr21_-_20328375 | 0.58 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr15_-_21132480 | 0.58 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr8_-_44298964 | 0.58 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr21_-_25748913 | 0.57 |
ENSDART00000134217
|
CR388166.2
|
|
| chr24_+_5237753 | 0.57 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr21_+_13301798 | 0.56 |
ENSDART00000140206
|
adora2ab
|
adenosine A2a receptor b |
| chr4_+_52356485 | 0.56 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr23_-_21446985 | 0.56 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr4_+_29165840 | 0.55 |
ENSDART00000169539
|
CR388132.2
|
|
| chr3_-_39488482 | 0.55 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr16_+_42830152 | 0.54 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr8_+_32755688 | 0.54 |
ENSDART00000137897
|
hmcn2
|
hemicentin 2 |
| chr14_+_26437891 | 0.54 |
ENSDART00000175459
|
gpr137
|
G protein-coupled receptor 137 |
| chr2_+_31838442 | 0.53 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr17_-_44249538 | 0.53 |
ENSDART00000008816
|
otx2b
|
orthodenticle homeobox 2b |
| chr3_+_24134418 | 0.53 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr12_+_27536095 | 0.53 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr3_-_39488639 | 0.52 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr4_+_5506952 | 0.52 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr3_+_62161184 | 0.52 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr1_-_15329277 | 0.52 |
ENSDART00000109970
|
dlc1
|
DLC1 Rho GTPase activating protein |
| chr13_+_17694845 | 0.52 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr25_+_13620555 | 0.51 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr2_+_47905735 | 0.50 |
ENSDART00000159701
|
ftr23
|
finTRIM family, member 23 |
| chr25_-_31423493 | 0.50 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr13_-_11986754 | 0.49 |
ENSDART00000164214
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr4_-_43280244 | 0.49 |
ENSDART00000150762
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr15_-_5901514 | 0.49 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr15_+_42431198 | 0.49 |
ENSDART00000189951
ENSDART00000089694 |
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr4_+_37406676 | 0.49 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr2_+_16781015 | 0.49 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr6_-_53426773 | 0.48 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr1_-_35924495 | 0.48 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr13_+_7442023 | 0.48 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr15_+_28685892 | 0.48 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr12_+_27537357 | 0.47 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr18_+_34599315 | 0.47 |
ENSDART00000159306
ENSDART00000088675 |
tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_-_32621103 | 0.47 |
ENSDART00000155321
|
xkr5b
|
XK related 5b |
| chr20_-_34127415 | 0.47 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr16_-_46573211 | 0.47 |
ENSDART00000193392
|
zgc:174938
|
zgc:174938 |
| chr14_+_34966598 | 0.46 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr24_-_33756003 | 0.46 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr21_+_11503212 | 0.46 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr12_+_25600685 | 0.46 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr1_+_47486104 | 0.46 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr12_+_19356623 | 0.46 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr7_-_24364536 | 0.45 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr11_+_7580079 | 0.45 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr2_+_16780643 | 0.45 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr22_+_18188045 | 0.45 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr17_-_23412705 | 0.44 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr10_-_10864331 | 0.44 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr9_+_45839260 | 0.44 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr4_-_70488123 | 0.43 |
ENSDART00000169266
|
si:dkeyp-44b5.5
|
si:dkeyp-44b5.5 |
| chr7_-_26076970 | 0.43 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr6_-_45869127 | 0.43 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr16_-_15387459 | 0.43 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr5_-_30715225 | 0.43 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr18_+_31073265 | 0.43 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr9_+_38158570 | 0.43 |
ENSDART00000059549
ENSDART00000133060 |
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr6_+_16468776 | 0.43 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr2_-_44283554 | 0.43 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr2_-_30659222 | 0.42 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr13_+_28675686 | 0.42 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr2_-_59145027 | 0.42 |
ENSDART00000128320
|
FO834803.1
|
|
| chr4_+_37239915 | 0.42 |
ENSDART00000146015
|
znf973
|
zinc finger protein 973 |
| chr13_+_22295905 | 0.42 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr9_+_32301017 | 0.42 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr22_+_18187857 | 0.42 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr7_+_49664174 | 0.42 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr5_-_33281046 | 0.41 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr4_-_5652030 | 0.41 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr6_+_58915889 | 0.41 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr16_+_42829735 | 0.40 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr21_-_22543611 | 0.40 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr3_+_31093455 | 0.40 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr2_-_30055432 | 0.40 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr15_+_46313082 | 0.40 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr8_+_14381272 | 0.39 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr8_-_13985032 | 0.39 |
ENSDART00000140576
|
igf3
|
insulin-like growth factor 3 |
| chr13_+_29519115 | 0.39 |
ENSDART00000086711
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr2_+_19195841 | 0.39 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr15_+_11814969 | 0.39 |
ENSDART00000127248
|
FO704748.1
|
|
| chr10_-_7988396 | 0.39 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr3_+_24275766 | 0.38 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr16_+_29509133 | 0.38 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_-_2310064 | 0.38 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr10_-_5016997 | 0.38 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr4_-_56898328 | 0.38 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr17_+_21295132 | 0.38 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr5_-_36948586 | 0.38 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr4_-_37707809 | 0.37 |
ENSDART00000162606
|
znf1139
|
zinc finger protein 1139 |
| chr11_+_30310170 | 0.37 |
ENSDART00000127797
|
ugt1b3
|
UDP glucuronosyltransferase 1 family, polypeptide B3 |
| chr20_-_29499363 | 0.37 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr12_+_19199735 | 0.37 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr4_+_73672430 | 0.37 |
ENSDART00000174310
ENSDART00000150505 |
si:dkey-262g12.7
|
si:dkey-262g12.7 |
| chr8_-_19216180 | 0.37 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr22_-_4644484 | 0.36 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr1_+_54683655 | 0.36 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr11_-_36279602 | 0.36 |
ENSDART00000122531
ENSDART00000103064 ENSDART00000125616 |
nfya
|
nuclear transcription factor Y, alpha |
| chr23_+_19813677 | 0.36 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr5_-_13685047 | 0.36 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr5_+_6955900 | 0.36 |
ENSDART00000099417
|
CABZ01041962.1
|
|
| chr14_+_15495088 | 0.36 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr23_+_43718115 | 0.36 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr6_+_43015916 | 0.36 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr4_-_45301719 | 0.35 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr20_-_14012859 | 0.35 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr4_+_63818212 | 0.35 |
ENSDART00000164929
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr5_-_40510397 | 0.35 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr7_-_6680622 | 0.35 |
ENSDART00000180437
ENSDART00000173270 ENSDART00000066450 |
dusp19b
|
dual specificity phosphatase 19b |
| chr8_+_30709685 | 0.35 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr1_+_5402476 | 0.34 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr4_+_45610254 | 0.34 |
ENSDART00000150866
|
znf1038
|
zinc finger protein 1038 |
| chr9_+_17306162 | 0.34 |
ENSDART00000075926
|
scel
|
sciellin |
| chr21_+_25236297 | 0.34 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr8_-_19216657 | 0.34 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr2_+_49569017 | 0.34 |
ENSDART00000109471
|
sema4e
|
semaphorin 4e |
| chr2_-_30135446 | 0.33 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr19_-_47571456 | 0.33 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr16_-_7793457 | 0.33 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr18_+_20034023 | 0.33 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr21_+_30351256 | 0.33 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr8_+_30671060 | 0.33 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr6_+_7977611 | 0.33 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr21_-_25756119 | 0.33 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr7_-_45999333 | 0.33 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr22_-_622937 | 0.33 |
ENSDART00000145685
ENSDART00000144309 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr6_-_57485411 | 0.33 |
ENSDART00000011663
|
snrpb
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr4_-_52621665 | 0.33 |
ENSDART00000137064
|
si:dkeyp-104f11.6
|
si:dkeyp-104f11.6 |
| chr4_+_13452907 | 0.33 |
ENSDART00000186116
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr9_+_32301456 | 0.33 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr9_-_32300783 | 0.32 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr8_-_25002182 | 0.32 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr2_-_24962820 | 0.31 |
ENSDART00000182767
|
hltf
|
helicase-like transcription factor |
| chr19_+_42469058 | 0.31 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr18_-_8380090 | 0.31 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr4_+_45148652 | 0.31 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr1_-_26664840 | 0.31 |
ENSDART00000102500
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr13_-_37619159 | 0.30 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr20_+_32501748 | 0.30 |
ENSDART00000152944
ENSDART00000021035 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr23_-_43718067 | 0.30 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr18_-_35407695 | 0.30 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr8_+_32747612 | 0.30 |
ENSDART00000142824
|
hmcn2
|
hemicentin 2 |
| chr22_+_465269 | 0.30 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr1_+_14466860 | 0.30 |
ENSDART00000114097
|
stim2a
|
tromal interaction molecule 2a |
| chr15_+_28685625 | 0.30 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr4_-_35899188 | 0.30 |
ENSDART00000167093
|
zgc:174653
|
zgc:174653 |
| chr21_-_11970199 | 0.29 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr8_-_13641022 | 0.29 |
ENSDART00000080875
|
b3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr17_-_49481672 | 0.29 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr8_+_14886452 | 0.29 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr5_+_32845759 | 0.29 |
ENSDART00000136552
|
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr18_+_38885309 | 0.29 |
ENSDART00000041597
|
arpp19a
|
cAMP-regulated phosphoprotein 19a |
| chr4_-_30440116 | 0.29 |
ENSDART00000159935
|
si:dkey-199m13.5
|
si:dkey-199m13.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa13a+hoxa13b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.5 | 1.4 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 1.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 0.7 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.5 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.2 | 0.6 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.2 | 0.6 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 0.4 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.8 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.8 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.6 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.4 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 1.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.1 | 3.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 1.5 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.2 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.3 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.2 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.6 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.2 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 1.7 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:1902746 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.2 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.6 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.1 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.2 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.1 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.3 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.0 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.4 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.0 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 3.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.7 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 1.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.2 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.0 | 0.2 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.2 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0017050 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |