Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
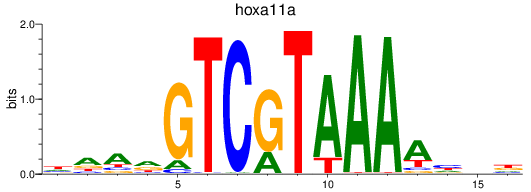
Results for hoxa11a
Z-value: 0.37
Transcription factors associated with hoxa11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11a
|
ENSDARG00000104162 | homeobox A11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11a | dr11_v1_chr19_+_19737214_19737300 | 0.30 | 4.3e-01 | Click! |
Activity profile of hoxa11a motif
Sorted Z-values of hoxa11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_39005317 | 0.46 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr7_+_20524064 | 0.46 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr12_-_26851726 | 0.43 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr12_+_27117609 | 0.40 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr11_+_2198831 | 0.31 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr22_+_10646928 | 0.30 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr2_-_10338759 | 0.29 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr22_-_10502780 | 0.28 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr9_-_3671911 | 0.25 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr17_-_31695217 | 0.23 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr15_+_36115955 | 0.23 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr7_+_67467702 | 0.22 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr1_+_31019165 | 0.22 |
ENSDART00000102210
|
itga6b
|
integrin, alpha 6b |
| chr23_+_36087219 | 0.22 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr3_+_36284986 | 0.21 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr16_+_20910186 | 0.20 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr7_-_43840418 | 0.19 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr17_+_42274825 | 0.19 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr23_+_27068225 | 0.19 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr1_-_462165 | 0.19 |
ENSDART00000152799
|
si:ch73-244f7.3
|
si:ch73-244f7.3 |
| chr2_-_3158919 | 0.19 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr24_+_21540842 | 0.16 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr15_+_8767650 | 0.14 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr10_+_33990572 | 0.13 |
ENSDART00000138547
|
fryb
|
furry homolog b (Drosophila) |
| chr20_-_46541834 | 0.12 |
ENSDART00000060685
ENSDART00000181720 |
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr7_+_31879649 | 0.12 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_24190207 | 0.12 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr7_+_31879986 | 0.11 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr5_-_35301800 | 0.11 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr20_+_43379029 | 0.11 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr16_-_10316359 | 0.11 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr1_+_45056371 | 0.10 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr3_+_59851537 | 0.10 |
ENSDART00000180997
|
CU693479.1
|
|
| chr15_-_14552101 | 0.10 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_-_25618175 | 0.09 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr10_+_2587234 | 0.09 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr11_-_36051004 | 0.09 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr19_-_22507715 | 0.09 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr4_-_9586713 | 0.08 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr11_+_24716837 | 0.08 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr18_-_2433011 | 0.08 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr4_+_39368978 | 0.08 |
ENSDART00000160640
|
si:dkey-261o4.1
|
si:dkey-261o4.1 |
| chr2_-_37874647 | 0.07 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr9_-_23922011 | 0.07 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr6_-_40744720 | 0.07 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr23_-_33750135 | 0.07 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr1_-_56213723 | 0.07 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr11_+_41135055 | 0.06 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr7_-_30280934 | 0.06 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr16_-_6205790 | 0.06 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr20_+_33532296 | 0.06 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr20_-_37820939 | 0.06 |
ENSDART00000032978
|
nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr14_-_48348973 | 0.06 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr23_-_1658980 | 0.06 |
ENSDART00000186227
|
CU693481.1
|
|
| chr2_-_9259283 | 0.05 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr2_+_43851983 | 0.05 |
ENSDART00000126413
|
nlrb5
|
NOD-like receptor family B, member 5 |
| chr21_+_13128180 | 0.05 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr7_+_20966434 | 0.05 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr21_+_13127742 | 0.05 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr23_-_33750307 | 0.04 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr12_-_34713690 | 0.04 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr22_+_2239974 | 0.04 |
ENSDART00000141993
|
znf1144
|
zinc finger protein 1144 |
| chr15_+_9297340 | 0.04 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr17_-_23727978 | 0.04 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr22_-_33916620 | 0.03 |
ENSDART00000191276
|
CR974456.1
|
|
| chr4_-_20108833 | 0.03 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr7_-_41014773 | 0.03 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr7_+_25036188 | 0.03 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr15_+_37950556 | 0.02 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr24_-_31223232 | 0.02 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr2_-_36914281 | 0.02 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr7_-_18508815 | 0.02 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr18_+_40993369 | 0.02 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr5_+_32932357 | 0.02 |
ENSDART00000192397
|
lmo4a
|
LIM domain only 4a |
| chr18_+_40993196 | 0.01 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr16_-_47426482 | 0.01 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr12_-_34713888 | 0.01 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr19_-_33370271 | 0.00 |
ENSDART00000132628
|
nkd3l
|
naked cuticle homolog 3, like |
| chr13_-_9875538 | 0.00 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr16_-_24561354 | 0.00 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |