Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
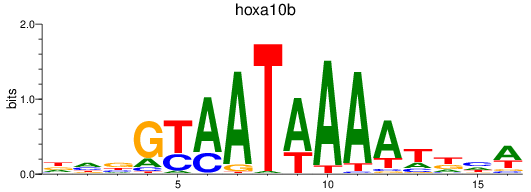
Results for hoxa10b
Z-value: 0.60
Transcription factors associated with hoxa10b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa10b
|
ENSDARG00000031337 | homeobox A10b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa10b | dr11_v1_chr16_+_20910186_20910186 | -0.88 | 1.6e-03 | Click! |
Activity profile of hoxa10b motif
Sorted Z-values of hoxa10b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_9485939 | 1.21 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr10_-_21545091 | 1.17 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr22_+_5574952 | 0.92 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr11_-_25257045 | 0.92 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr7_+_1473929 | 0.90 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr15_-_43978141 | 0.84 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr24_-_21090447 | 0.78 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr17_+_43595692 | 0.72 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr2_+_35733335 | 0.69 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr1_-_53468160 | 0.69 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr17_+_50701748 | 0.69 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr12_-_10512911 | 0.68 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr13_+_8892784 | 0.68 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr16_-_27566552 | 0.67 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr13_+_33462232 | 0.67 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr2_-_17115256 | 0.62 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_-_50062909 | 0.62 |
ENSDART00000075209
|
zgc:113886
|
zgc:113886 |
| chr14_-_7409364 | 0.62 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr16_-_44945224 | 0.61 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr5_-_31856681 | 0.61 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr10_+_40321067 | 0.60 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr17_-_30652738 | 0.60 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr13_-_25719628 | 0.60 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr21_+_43178831 | 0.59 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr5_-_57723929 | 0.58 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr5_-_37881345 | 0.58 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr3_-_29910547 | 0.58 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr5_-_57289872 | 0.58 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr12_-_18961289 | 0.57 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr11_+_11303458 | 0.57 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr25_+_9013342 | 0.56 |
ENSDART00000154207
ENSDART00000153705 |
im:7145024
|
im:7145024 |
| chr14_+_45028062 | 0.56 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr23_+_28092083 | 0.55 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr13_-_17860307 | 0.53 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr10_+_6013076 | 0.52 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr1_+_2321384 | 0.51 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr24_+_10202718 | 0.51 |
ENSDART00000126668
|
pou6f2
|
POU class 6 homeobox 2 |
| chr9_+_34232503 | 0.51 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr5_-_31857345 | 0.50 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr6_-_9282080 | 0.50 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr25_+_3549841 | 0.49 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr20_-_27190393 | 0.49 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr11_+_31730680 | 0.47 |
ENSDART00000145497
|
diaph3
|
diaphanous-related formin 3 |
| chr22_+_23430688 | 0.46 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr24_-_23784701 | 0.46 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr14_-_50028428 | 0.45 |
ENSDART00000163543
ENSDART00000168510 |
abhd18
|
abhydrolase domain containing 18 |
| chr12_-_18961491 | 0.45 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr2_-_10877765 | 0.44 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr2_+_49457449 | 0.44 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr10_+_15340768 | 0.44 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr12_-_4243268 | 0.44 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr10_-_20588787 | 0.43 |
ENSDART00000138045
ENSDART00000181885 ENSDART00000091115 |
nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr8_+_2656231 | 0.43 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr18_-_39288894 | 0.43 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr21_-_14830353 | 0.43 |
ENSDART00000134687
|
pus1
|
pseudouridylate synthase 1 |
| chr13_-_27354003 | 0.42 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr22_+_15959844 | 0.42 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr15_+_29727799 | 0.42 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr1_-_14506759 | 0.41 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr23_+_45200481 | 0.40 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr5_+_20453874 | 0.40 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr15_+_22390076 | 0.40 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr5_-_23715861 | 0.40 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr13_-_24717365 | 0.40 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr1_-_8566567 | 0.39 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr11_-_25257595 | 0.39 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr21_+_19334198 | 0.39 |
ENSDART00000147372
|
helq
|
helicase, POLQ like |
| chr13_+_15580758 | 0.39 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr6_-_7107868 | 0.39 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr13_-_24717618 | 0.38 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr13_+_13945218 | 0.38 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr20_-_29499363 | 0.38 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr2_+_51818039 | 0.38 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr24_-_15159658 | 0.38 |
ENSDART00000142473
|
rttn
|
rotatin |
| chr2_-_40889465 | 0.38 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr11_+_6010177 | 0.38 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr11_-_26362294 | 0.38 |
ENSDART00000184654
ENSDART00000115037 |
foxj3
|
forkhead box J3 |
| chr18_-_7097403 | 0.37 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr3_-_60589292 | 0.36 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr10_-_31805923 | 0.36 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr19_+_40069524 | 0.36 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr6_+_40554551 | 0.36 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr1_-_55068941 | 0.35 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr14_-_899170 | 0.35 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr1_-_41374117 | 0.35 |
ENSDART00000074777
|
htt
|
huntingtin |
| chr22_-_12337781 | 0.35 |
ENSDART00000188357
ENSDART00000123574 |
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr22_+_2512154 | 0.35 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr15_-_1843831 | 0.34 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_40890004 | 0.34 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_+_41596099 | 0.34 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr21_+_4313039 | 0.34 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr23_+_20431140 | 0.33 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr5_-_65121747 | 0.33 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr2_-_37458527 | 0.33 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr2_+_49805892 | 0.33 |
ENSDART00000056248
|
wdr48b
|
WD repeat domain 48b |
| chr22_+_1953096 | 0.33 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr2_-_20715094 | 0.33 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr23_-_24253200 | 0.33 |
ENSDART00000114840
|
zbtb17
|
zinc finger and BTB domain containing 17 |
| chr20_-_2641233 | 0.32 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr18_+_21951551 | 0.32 |
ENSDART00000146261
|
ranbp10
|
RAN binding protein 10 |
| chr12_+_32368574 | 0.32 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr7_+_15308219 | 0.32 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr22_-_24818066 | 0.32 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr25_-_29074064 | 0.32 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr25_+_16080181 | 0.31 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr16_+_46401756 | 0.31 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr1_-_12397258 | 0.31 |
ENSDART00000144596
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr7_+_55149001 | 0.31 |
ENSDART00000148642
|
cdh31
|
cadherin 31 |
| chr19_+_7424347 | 0.31 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr18_+_15876385 | 0.31 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr5_-_16405651 | 0.30 |
ENSDART00000163942
|
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr10_-_15340362 | 0.30 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr20_-_34768301 | 0.30 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr19_-_47571797 | 0.30 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_-_8892514 | 0.30 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr14_-_238931 | 0.30 |
ENSDART00000161122
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr2_+_22409249 | 0.30 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr11_-_25461336 | 0.30 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr8_-_31416403 | 0.29 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr22_+_10606573 | 0.29 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr17_-_25630822 | 0.29 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr12_-_6551681 | 0.29 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr1_+_12394205 | 0.29 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr13_+_7575563 | 0.28 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr13_+_33268657 | 0.28 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr13_+_1131748 | 0.28 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr17_-_31212420 | 0.28 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr8_-_39952727 | 0.28 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr15_-_1733235 | 0.27 |
ENSDART00000023153
ENSDART00000154668 |
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr2_+_54086436 | 0.27 |
ENSDART00000174581
|
CU179656.1
|
|
| chr17_+_17764979 | 0.27 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr12_+_41348969 | 0.27 |
ENSDART00000171352
|
si:ch211-27e6.1
|
si:ch211-27e6.1 |
| chr1_+_57371447 | 0.27 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr21_-_14878220 | 0.27 |
ENSDART00000131237
|
ulk1b
|
unc-51 like autophagy activating kinase 1 |
| chr7_+_17907883 | 0.27 |
ENSDART00000147564
|
mta2
|
metastasis associated 1 family, member 2 |
| chr1_-_49947482 | 0.27 |
ENSDART00000143398
|
sgms2
|
sphingomyelin synthase 2 |
| chr12_+_33361948 | 0.27 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr16_-_32671998 | 0.27 |
ENSDART00000161395
|
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr16_+_36126310 | 0.26 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr12_+_11352630 | 0.26 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr19_-_47571456 | 0.26 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_+_7279104 | 0.26 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr7_-_7692723 | 0.25 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr23_-_36823932 | 0.25 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr14_+_10625112 | 0.25 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr8_-_10947205 | 0.25 |
ENSDART00000164467
|
pqlc2
|
PQ loop repeat containing 2 |
| chr12_+_23912074 | 0.25 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr4_-_11053543 | 0.25 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr8_-_18899427 | 0.24 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr12_-_11560794 | 0.24 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr10_-_11840353 | 0.24 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr13_+_29292011 | 0.24 |
ENSDART00000115023
|
parga
|
poly (ADP-ribose) glycohydrolase a |
| chr6_+_45692026 | 0.24 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr10_+_2975974 | 0.24 |
ENSDART00000147918
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr25_-_27633115 | 0.23 |
ENSDART00000131539
|
hyal6
|
hyaluronoglucosaminidase 6 |
| chr11_+_12052791 | 0.23 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr4_+_23125689 | 0.23 |
ENSDART00000077854
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr10_-_33297864 | 0.23 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr12_-_16452200 | 0.23 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr13_+_24717880 | 0.23 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr16_+_20934353 | 0.22 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr3_+_60589157 | 0.22 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr2_-_49627099 | 0.22 |
ENSDART00000083710
ENSDART00000147421 |
cactin
|
cactin |
| chr17_+_21760032 | 0.22 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr3_+_31093455 | 0.22 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr23_+_43718115 | 0.22 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr22_+_30195257 | 0.22 |
ENSDART00000027803
ENSDART00000172496 |
add3a
|
adducin 3 (gamma) a |
| chr24_+_14581864 | 0.22 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr1_-_13989643 | 0.22 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr5_+_24063046 | 0.22 |
ENSDART00000051548
ENSDART00000133355 ENSDART00000142268 |
gps2
|
G protein pathway suppressor 2 |
| chr2_-_50272077 | 0.22 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr2_+_30960351 | 0.21 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr25_+_3549401 | 0.21 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr13_+_9559461 | 0.21 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr23_-_14766902 | 0.21 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr5_+_27137473 | 0.21 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr13_-_9070754 | 0.21 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr4_-_17353972 | 0.21 |
ENSDART00000041529
|
parpbp
|
PARP1 binding protein |
| chr16_+_31942527 | 0.20 |
ENSDART00000188334
ENSDART00000188643 ENSDART00000058496 |
phc1
|
polyhomeotic homolog 1 |
| chr5_+_50913034 | 0.20 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr13_+_9468535 | 0.20 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr3_+_26135502 | 0.20 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr11_-_5953636 | 0.20 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr21_-_3007412 | 0.19 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr7_-_16582187 | 0.19 |
ENSDART00000131726
|
e2f8
|
E2F transcription factor 8 |
| chr15_-_23908605 | 0.19 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr22_-_20950448 | 0.19 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr15_-_26931541 | 0.19 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr2_-_30912922 | 0.19 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr25_+_22571475 | 0.19 |
ENSDART00000161559
|
stra6
|
stimulated by retinoic acid 6 |
| chr12_-_10476448 | 0.18 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr8_+_39795918 | 0.18 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr3_+_39579393 | 0.18 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr8_-_11324143 | 0.18 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr19_+_27970292 | 0.18 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr15_-_23908779 | 0.18 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr20_+_32552912 | 0.17 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr7_+_22691961 | 0.17 |
ENSDART00000041615
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr14_+_22467672 | 0.17 |
ENSDART00000079409
ENSDART00000136597 |
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr16_+_38337783 | 0.17 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa10b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.5 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.6 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.3 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.4 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.8 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.2 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.8 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.3 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0007440 | foregut morphogenesis(GO:0007440) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.5 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.5 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.7 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.6 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 1.1 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |