Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
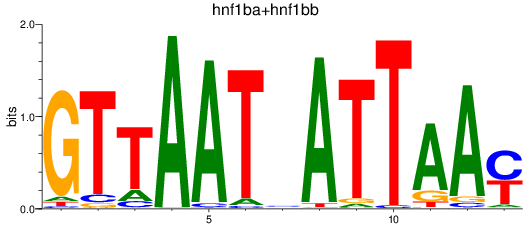
Results for hnf1ba+hnf1bb
Z-value: 2.39
Transcription factors associated with hnf1ba+hnf1bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1ba
|
ENSDARG00000006615 | HNF1 homeobox Ba |
|
hnf1bb
|
ENSDARG00000022295 | HNF1 homeobox Bb |
|
hnf1ba
|
ENSDARG00000115522 | HNF1 homeobox Ba |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1ba | dr11_v1_chr15_-_16012963_16012963 | 0.60 | 9.0e-02 | Click! |
| hnf1bb | dr11_v1_chr21_+_38817785_38817843 | 0.52 | 1.5e-01 | Click! |
Activity profile of hnf1ba+hnf1bb motif
Sorted Z-values of hnf1ba+hnf1bb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_32345187 | 6.87 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr2_+_55984788 | 5.31 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr22_-_23668356 | 5.11 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr6_+_60055168 | 4.73 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr15_+_28202170 | 4.70 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr20_+_10538025 | 4.67 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr1_+_10051763 | 4.43 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr19_+_19772765 | 4.37 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr24_-_39610585 | 4.29 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr16_-_42894628 | 4.26 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr7_-_25895189 | 4.22 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr22_-_23748284 | 4.21 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr20_+_10544100 | 3.80 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr5_+_37854685 | 3.72 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr5_-_19394440 | 3.69 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr2_+_1487118 | 3.57 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr1_-_12126535 | 3.55 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr8_+_15254564 | 3.52 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr16_+_26777473 | 3.37 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr23_-_20345473 | 3.35 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr8_-_1051438 | 3.33 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr18_-_48530221 | 3.19 |
ENSDART00000188134
ENSDART00000142107 |
kcnj1a.2
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 2 |
| chr2_+_1486822 | 3.09 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr24_+_14713776 | 2.87 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr5_-_41307550 | 2.83 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr25_-_27541621 | 2.72 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr4_+_7827261 | 2.72 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr23_+_32335871 | 2.66 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr3_+_40170216 | 2.61 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr13_-_40726865 | 2.56 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr9_+_21151138 | 2.53 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr2_-_17828279 | 2.50 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr21_-_20328375 | 2.44 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr15_+_35043007 | 2.41 |
ENSDART00000086954
|
sesn3
|
sestrin 3 |
| chr10_+_25369254 | 2.39 |
ENSDART00000164375
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr6_+_7977611 | 2.39 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr21_+_43669943 | 2.33 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr22_-_23706771 | 2.33 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr12_-_3940768 | 2.30 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr18_-_48500545 | 2.28 |
ENSDART00000058989
ENSDART00000098450 |
kcnj1a.6
kcnj1a.5
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 5 |
| chr23_+_28770225 | 2.28 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr3_+_26734162 | 2.22 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr16_-_9675982 | 2.21 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr8_-_47800754 | 2.17 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr2_+_26647472 | 2.16 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr24_-_17039638 | 2.14 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr21_-_23308286 | 2.14 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr9_-_14108896 | 2.11 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr24_-_6078222 | 2.08 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr22_-_26100282 | 2.01 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr5_-_37886063 | 1.97 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr18_-_48550426 | 1.96 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr2_-_43635777 | 1.96 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr14_+_38654300 | 1.93 |
ENSDART00000148938
ENSDART00000150107 |
rnasel3
|
ribonuclease like 3 |
| chr22_-_26005894 | 1.88 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr12_-_6177894 | 1.87 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr13_+_23988442 | 1.86 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr2_-_5466708 | 1.82 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr8_-_18535822 | 1.81 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr7_-_4296771 | 1.81 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr6_+_54221654 | 1.77 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr17_+_15033822 | 1.68 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr24_+_23791758 | 1.66 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr25_+_3347461 | 1.63 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr8_+_26522013 | 1.62 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr13_-_36534380 | 1.60 |
ENSDART00000177857
ENSDART00000181046 ENSDART00000187243 |
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr10_-_41664427 | 1.59 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr17_-_17948587 | 1.59 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr18_-_48492951 | 1.58 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr10_+_35439952 | 1.58 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr24_-_4765740 | 1.58 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_+_45085194 | 1.57 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr9_+_29994439 | 1.56 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr2_-_5475910 | 1.50 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr6_+_30456788 | 1.50 |
ENSDART00000121492
|
FP236735.1
|
|
| chr1_-_22851481 | 1.45 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr13_-_18835254 | 1.43 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr8_-_53490376 | 1.42 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr10_-_17159761 | 1.42 |
ENSDART00000080449
|
slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr25_+_13406069 | 1.41 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr7_+_19817306 | 1.32 |
ENSDART00000044425
|
BX957278.1
|
|
| chr5_+_45322734 | 1.29 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr21_+_39100289 | 1.28 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr18_+_13182528 | 1.27 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr1_+_26667872 | 1.27 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr23_+_27740788 | 1.24 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr18_-_48508585 | 1.23 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr10_+_25369859 | 1.22 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr3_-_16250527 | 1.21 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr20_-_25481306 | 1.19 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr9_+_30720048 | 1.17 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr5_+_21225017 | 1.16 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr21_+_39941184 | 1.16 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr7_-_29534001 | 1.13 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr3_-_16760923 | 1.12 |
ENSDART00000055855
|
aspdh
|
aspartate dehydrogenase domain containing |
| chr6_+_28141135 | 1.11 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr19_+_8144556 | 1.09 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr21_+_39941559 | 1.07 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr7_-_59564011 | 1.06 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr11_+_575665 | 1.05 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr18_-_48517040 | 1.03 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr23_+_25856541 | 1.03 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr10_+_25726694 | 0.99 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr21_+_38002879 | 0.95 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr22_+_16555939 | 0.95 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_-_56103718 | 0.95 |
ENSDART00000191002
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr8_+_46418996 | 0.90 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr24_+_35787629 | 0.88 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr6_+_38381957 | 0.87 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr8_+_26293673 | 0.86 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr20_+_23440632 | 0.86 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr4_+_76416268 | 0.82 |
ENSDART00000174351
ENSDART00000191591 |
zgc:113209
FP074874.1
|
zgc:113209 |
| chr5_-_42904329 | 0.82 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr23_+_27740592 | 0.75 |
ENSDART00000137875
|
dhh
|
desert hedgehog |
| chr19_-_22568203 | 0.74 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr4_+_45484774 | 0.74 |
ENSDART00000150573
|
si:dkey-256i11.6
|
si:dkey-256i11.6 |
| chr24_+_25913162 | 0.74 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr4_-_9533345 | 0.71 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr5_-_65158203 | 0.67 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr4_-_73136420 | 0.66 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr1_-_19502322 | 0.65 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr10_+_21867307 | 0.64 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr4_+_58016732 | 0.63 |
ENSDART00000165777
|
CT027609.1
|
|
| chr5_-_63302944 | 0.62 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr24_-_21913426 | 0.60 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr14_+_740032 | 0.58 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr4_+_54798291 | 0.58 |
ENSDART00000165113
ENSDART00000109624 |
si:dkeyp-82b4.6
|
si:dkeyp-82b4.6 |
| chr8_+_30699429 | 0.54 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr16_-_22294265 | 0.53 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr16_+_11623956 | 0.51 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr15_+_25156382 | 0.51 |
ENSDART00000171092
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr25_+_23280220 | 0.49 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr15_+_29283981 | 0.49 |
ENSDART00000099895
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr3_+_28576173 | 0.49 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr3_-_34084387 | 0.48 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr2_-_41942666 | 0.46 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr19_-_18418763 | 0.44 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr4_+_34119824 | 0.44 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr4_-_56954002 | 0.44 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr5_-_30715225 | 0.44 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr20_-_31075972 | 0.43 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr7_-_27696958 | 0.42 |
ENSDART00000173470
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_+_28140816 | 0.42 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr4_-_67980261 | 0.41 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr7_-_35066457 | 0.41 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr4_-_32642681 | 0.40 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr12_+_5209822 | 0.40 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr4_+_18804317 | 0.38 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr16_+_11603096 | 0.37 |
ENSDART00000146286
|
si:dkey-11o1.3
|
si:dkey-11o1.3 |
| chr20_-_40755614 | 0.35 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr21_-_38853737 | 0.34 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr11_-_37997419 | 0.34 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr15_-_41744509 | 0.34 |
ENSDART00000176952
ENSDART00000156690 |
ftr88
|
finTRIM family, member 88 |
| chr3_+_19446997 | 0.32 |
ENSDART00000079352
|
zgc:123297
|
zgc:123297 |
| chr18_+_3530769 | 0.30 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr17_-_47305034 | 0.29 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr7_+_27059330 | 0.28 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr4_-_32179699 | 0.28 |
ENSDART00000124106
ENSDART00000158835 |
si:dkey-72l17.6
|
si:dkey-72l17.6 |
| chr25_+_17920361 | 0.27 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr5_+_13385837 | 0.25 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr21_-_23307653 | 0.25 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr17_+_5895500 | 0.24 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr4_-_5317006 | 0.24 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr2_-_7845110 | 0.23 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr15_+_25156121 | 0.23 |
ENSDART00000189709
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr18_+_2554666 | 0.22 |
ENSDART00000167218
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr20_-_40750953 | 0.20 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr23_+_26491931 | 0.20 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr15_+_35139457 | 0.18 |
ENSDART00000059204
|
slc19a3a
|
solute carrier family 19 (thiamine transporter), member 3a |
| chr25_-_3759322 | 0.13 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr2_-_27609189 | 0.12 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr7_+_27317174 | 0.11 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_-_17412559 | 0.11 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr4_-_34480599 | 0.11 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr22_+_21317597 | 0.09 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr8_+_23391263 | 0.05 |
ENSDART00000149687
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr5_+_1877464 | 0.04 |
ENSDART00000050658
|
zgc:101699
|
zgc:101699 |
| chr9_+_28688574 | 0.04 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr19_+_28256076 | 0.03 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr10_-_42131408 | 0.02 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr18_+_33353271 | 0.02 |
ENSDART00000099142
|
v2rh13
|
vomeronasal 2 receptor, h13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1ba+hnf1bb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.2 | 4.7 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.9 | 3.7 | GO:2000252 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.8 | 3.9 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.6 | 4.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.6 | 3.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.6 | 2.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.5 | 1.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.5 | 2.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 3.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.4 | 2.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.4 | 2.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 1.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 2.4 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.3 | 2.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.3 | 4.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 3.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.3 | 2.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 2.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 2.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.3 | 6.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 1.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 1.5 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 2.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 2.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 2.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 2.8 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.6 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 11.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 2.2 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.1 | 0.5 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.9 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 1.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.0 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 5.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.8 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 2.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 4.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.5 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.5 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 3.4 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.5 | 4.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 6.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 34.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 2.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.9 | 2.6 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.8 | 4.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 11.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.8 | 5.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.6 | 2.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 3.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 2.0 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 2.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.6 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.3 | 1.6 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 2.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 2.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 0.8 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.2 | 4.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 3.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.5 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 4.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 2.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 2.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 5.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 7.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.1 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 2.0 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 1.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.1 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 7.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 5.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.3 | 5.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.8 | 17.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 2.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 4.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 3.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 4.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.4 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |