Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
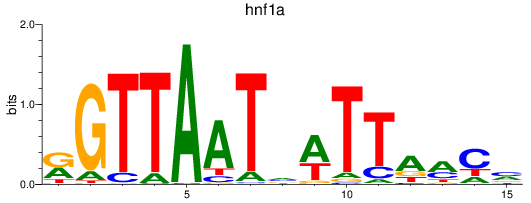
Results for hnf1a
Z-value: 1.76
Transcription factors associated with hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1a
|
ENSDARG00000009470 | HNF1 homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1a | dr11_v1_chr8_+_39802506_39802506 | -0.62 | 7.4e-02 | Click! |
Activity profile of hnf1a motif
Sorted Z-values of hnf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_45920 | 4.54 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr5_+_32345187 | 3.90 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr8_+_999421 | 3.72 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr22_-_23625351 | 3.66 |
ENSDART00000192588
ENSDART00000163423 |
cfhl4
|
complement factor H like 4 |
| chr24_-_39610585 | 3.30 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr17_+_4030493 | 3.19 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr22_-_23668356 | 3.11 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr16_-_42894628 | 2.85 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr1_+_10051763 | 2.83 |
ENSDART00000011701
|
fgb
|
fibrinogen beta chain |
| chr22_-_17595310 | 2.77 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr20_+_10544100 | 2.69 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr25_+_31323978 | 2.68 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr13_-_40726865 | 2.65 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr2_+_55984788 | 2.28 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr22_-_23748284 | 2.22 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr25_-_27541621 | 2.21 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr22_+_12770877 | 2.13 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr20_+_18741089 | 2.13 |
ENSDART00000183218
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr3_+_15505275 | 2.09 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr22_-_17611742 | 2.05 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr23_+_32335871 | 1.95 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr16_+_11724230 | 1.93 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr1_-_12126535 | 1.87 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr12_+_15666949 | 1.81 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr8_+_26522013 | 1.73 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr24_-_32665283 | 1.73 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr15_+_27364394 | 1.70 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr21_+_25236297 | 1.70 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr21_-_23308286 | 1.68 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr12_-_20373058 | 1.63 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr5_-_56513825 | 1.62 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr7_-_25895189 | 1.60 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr10_-_41664427 | 1.58 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr8_+_13106760 | 1.51 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr4_+_7827261 | 1.51 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr10_-_8033468 | 1.49 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr23_-_20345473 | 1.47 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr21_+_37442458 | 1.42 |
ENSDART00000180161
ENSDART00000186950 |
cox7b
|
cytochrome c oxidase subunit VIIb |
| chr5_-_37886063 | 1.42 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr11_+_43419809 | 1.41 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr6_+_7977611 | 1.38 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr22_-_26100282 | 1.37 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr24_-_17039638 | 1.37 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr2_-_17828279 | 1.36 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr21_-_44104600 | 1.35 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr7_-_4296771 | 1.35 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr9_+_21151138 | 1.33 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr9_-_14108896 | 1.31 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr24_+_14713776 | 1.31 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr11_-_23332592 | 1.29 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr12_-_6177894 | 1.29 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr12_+_31673588 | 1.28 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr12_-_6172154 | 1.27 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr8_-_18535822 | 1.27 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr5_+_26795773 | 1.26 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr22_-_26005894 | 1.26 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr24_-_4765740 | 1.23 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_+_26647472 | 1.23 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr16_+_55059026 | 1.23 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr25_-_27541288 | 1.22 |
ENSDART00000187245
|
spam1
|
sperm adhesion molecule 1 |
| chr22_-_23706771 | 1.22 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr7_+_27250186 | 1.21 |
ENSDART00000150068
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr10_+_20108557 | 1.20 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr8_-_47800754 | 1.11 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr5_+_9348284 | 1.09 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr3_+_49521106 | 1.09 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr3_+_40170216 | 1.08 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr2_-_43635777 | 1.07 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr8_-_13013123 | 1.05 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr10_+_4499943 | 1.02 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr16_-_9675982 | 1.02 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr10_-_8032885 | 1.00 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr20_+_22666548 | 0.99 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr10_+_35439952 | 0.98 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr21_+_44684577 | 0.96 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr7_-_59564011 | 0.94 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr15_-_23467750 | 0.93 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr1_-_22851481 | 0.92 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr21_+_43669943 | 0.90 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr18_+_13182528 | 0.90 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr20_+_18740799 | 0.89 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr6_+_30456788 | 0.87 |
ENSDART00000121492
|
FP236735.1
|
|
| chr6_+_34028532 | 0.87 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr24_+_23791758 | 0.86 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr13_+_23988442 | 0.86 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_+_26734162 | 0.86 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr2_-_5475910 | 0.83 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr3_-_16250527 | 0.83 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr6_+_54221654 | 0.83 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr1_-_21409877 | 0.81 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr9_+_4429593 | 0.81 |
ENSDART00000184855
|
FP015810.1
|
|
| chr22_-_36770649 | 0.77 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr2_-_5466708 | 0.74 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr1_+_26667872 | 0.73 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr17_+_15033822 | 0.71 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr19_-_22387141 | 0.71 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr7_-_35066457 | 0.71 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr13_-_42579437 | 0.71 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| chr15_-_17869115 | 0.71 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr8_+_7144066 | 0.69 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr24_-_20016817 | 0.68 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr19_+_24882845 | 0.67 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr23_-_24394719 | 0.67 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr15_-_17868870 | 0.67 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr7_-_29625509 | 0.66 |
ENSDART00000173723
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr2_-_5404466 | 0.66 |
ENSDART00000152907
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr9_+_30720048 | 0.63 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr5_+_45322734 | 0.60 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr21_-_23307653 | 0.60 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr14_-_32884138 | 0.60 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr2_+_49417900 | 0.59 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr24_-_23675446 | 0.58 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr22_+_38978084 | 0.56 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr4_+_57881965 | 0.56 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr20_-_7293837 | 0.54 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr8_+_1187928 | 0.54 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr7_-_10560964 | 0.54 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr9_+_29994439 | 0.54 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr18_-_5111449 | 0.52 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr20_-_25481306 | 0.52 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr6_+_20647155 | 0.51 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr20_+_18740518 | 0.51 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr15_+_9297340 | 0.46 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr4_+_2620751 | 0.46 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr5_+_21225017 | 0.44 |
ENSDART00000141573
|
si:dkey-13n15.11
|
si:dkey-13n15.11 |
| chr11_+_25129013 | 0.40 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr3_-_34337969 | 0.40 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr10_+_21867307 | 0.39 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr21_+_39100289 | 0.39 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_+_31039923 | 0.38 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr5_-_63302944 | 0.38 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr21_+_38002879 | 0.37 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr4_-_9533345 | 0.37 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr14_-_17622080 | 0.37 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr6_+_8622228 | 0.36 |
ENSDART00000110470
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr6_-_39344259 | 0.36 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr7_-_29534001 | 0.35 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr6_+_28141135 | 0.35 |
ENSDART00000113405
ENSDART00000179795 |
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr15_+_29283981 | 0.33 |
ENSDART00000099895
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr20_+_35473288 | 0.32 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr14_+_740032 | 0.32 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr1_-_59348118 | 0.32 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr1_-_19502322 | 0.32 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr20_+_6543674 | 0.31 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr9_-_9977827 | 0.31 |
ENSDART00000187315
ENSDART00000010246 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr5_+_9377005 | 0.30 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr23_+_25856541 | 0.30 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr4_+_18804317 | 0.30 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr19_-_22568203 | 0.29 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr16_+_11623956 | 0.29 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr3_+_54761569 | 0.29 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr20_-_25436389 | 0.27 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr16_+_11603096 | 0.26 |
ENSDART00000146286
|
si:dkey-11o1.3
|
si:dkey-11o1.3 |
| chr4_+_76416268 | 0.26 |
ENSDART00000174351
ENSDART00000191591 |
zgc:113209
FP074874.1
|
zgc:113209 |
| chr5_-_29488245 | 0.26 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr9_-_9982696 | 0.25 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr4_-_56954002 | 0.25 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr5_+_9382301 | 0.25 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr18_+_9382847 | 0.24 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr3_-_34084387 | 0.24 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr4_+_34119824 | 0.24 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr12_+_6117682 | 0.23 |
ENSDART00000146650
|
asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr6_+_38381957 | 0.23 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr5_+_9417409 | 0.22 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr25_+_23280220 | 0.21 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr6_-_56103718 | 0.19 |
ENSDART00000191002
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr4_+_45484774 | 0.19 |
ENSDART00000150573
|
si:dkey-256i11.6
|
si:dkey-256i11.6 |
| chr18_+_15683157 | 0.19 |
ENSDART00000140933
|
si:ch211-264e16.2
|
si:ch211-264e16.2 |
| chr3_+_28576173 | 0.19 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr2_-_41942666 | 0.18 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr21_+_21803596 | 0.18 |
ENSDART00000084600
ENSDART00000184899 |
neu3.3
|
sialidase 3 (membrane sialidase), tandem duplicate 3 |
| chr19_+_20274491 | 0.18 |
ENSDART00000090860
ENSDART00000151693 |
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr2_-_48396061 | 0.17 |
ENSDART00000161657
|
si:ch211-195j11.30
|
si:ch211-195j11.30 |
| chr17_-_47305034 | 0.17 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr21_-_31286428 | 0.16 |
ENSDART00000185334
|
ca4c
|
carbonic anhydrase IV c |
| chr14_-_11507211 | 0.16 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr5_-_65158203 | 0.15 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr9_-_1459985 | 0.15 |
ENSDART00000124777
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr19_+_20274944 | 0.14 |
ENSDART00000151237
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr2_-_24402341 | 0.12 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr21_-_38853737 | 0.11 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr4_-_32642681 | 0.11 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr2_-_44746723 | 0.10 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr5_+_13385837 | 0.10 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr21_+_21803363 | 0.10 |
ENSDART00000081184
|
neu3.3
|
sialidase 3 (membrane sialidase), tandem duplicate 3 |
| chr9_+_27339505 | 0.09 |
ENSDART00000011822
|
slc10a2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr12_+_5209822 | 0.09 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr6_+_28140816 | 0.09 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr11_-_37997419 | 0.08 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr2_-_27609189 | 0.06 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr10_+_7255695 | 0.05 |
ENSDART00000161506
ENSDART00000181304 |
fut10
|
fucosyltransferase 10 |
| chr9_+_28688574 | 0.05 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr4_-_67980261 | 0.05 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr16_-_31980948 | 0.05 |
ENSDART00000182427
|
CR854927.4
|
|
| chr18_+_17786548 | 0.04 |
ENSDART00000189493
ENSDART00000146133 |
ZNF423
|
si:ch211-216l23.1 |
| chr19_+_31043002 | 0.04 |
ENSDART00000171591
|
ankmy2b
|
ankyrin repeat and MYND domain containing 2b |
| chr18_+_3530769 | 0.04 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr3_-_18047150 | 0.04 |
ENSDART00000020376
|
hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr15_+_35139457 | 0.03 |
ENSDART00000059204
|
slc19a3a
|
solute carrier family 19 (thiamine transporter), member 3a |
| chr5_+_9405283 | 0.03 |
ENSDART00000127306
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.1 | 3.3 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.7 | 2.1 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.7 | 4.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.6 | 1.8 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.5 | 2.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.4 | 1.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 1.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.3 | 1.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.3 | 4.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 3.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.4 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 1.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 1.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 1.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 2.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 2.6 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.2 | 1.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 2.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 2.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 0.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 0.5 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 1.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.6 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.2 | GO:0006775 | fat-soluble vitamin metabolic process(GO:0006775) |
| 0.1 | 0.4 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 1.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 0.9 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 4.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 1.4 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.8 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 1.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 1.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0044218 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.9 | 2.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 3.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.5 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.2 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.9 | 2.7 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.6 | 1.8 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.5 | 2.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.5 | 1.9 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 4.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 2.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 0.9 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.3 | 2.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.6 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 3.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 3.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 1.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 4.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 2.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.5 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 2.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.4 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.8 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 1.0 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 5.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 5.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |