Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
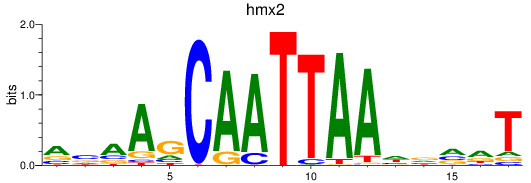
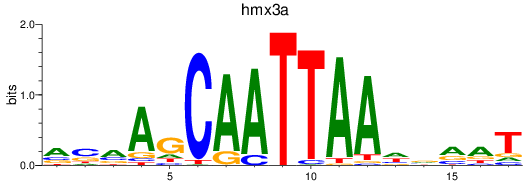
Results for hmx2_hmx3a
Z-value: 1.79
Transcription factors associated with hmx2_hmx3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx2
|
ENSDARG00000070954 | H6 family homeobox 2 |
|
hmx2
|
ENSDARG00000115364 | H6 family homeobox 2 |
|
hmx3a
|
ENSDARG00000070955 | H6 family homeobox 3a |
|
hmx3a
|
ENSDARG00000115051 | H6 family homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx2 | dr11_v1_chr17_-_21784152_21784152 | -0.95 | 1.1e-04 | Click! |
| hmx3a | dr11_v1_chr17_-_21793113_21793113 | -0.92 | 4.4e-04 | Click! |
Activity profile of hmx2_hmx3a motif
Sorted Z-values of hmx2_hmx3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_9997948 | 3.45 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_10014512 | 3.43 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr17_-_27223965 | 2.67 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr11_+_18157260 | 2.66 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr17_+_50701748 | 2.55 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr10_-_1961930 | 2.54 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr11_+_18130300 | 2.51 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr7_-_4461104 | 2.21 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr19_-_868187 | 2.18 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr15_-_41689981 | 2.08 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr16_+_28881235 | 1.81 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr23_+_45845159 | 1.68 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr11_+_25539698 | 1.66 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr5_-_65662996 | 1.59 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr19_+_41479990 | 1.57 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr19_-_34873566 | 1.54 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr19_-_7043355 | 1.50 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr22_-_20695237 | 1.47 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr2_+_56657804 | 1.44 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr15_-_33172246 | 1.40 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr1_+_15258641 | 1.37 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr8_+_41035230 | 1.36 |
ENSDART00000141349
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr24_-_38083378 | 1.36 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr18_-_26715655 | 1.35 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr16_+_40043673 | 1.35 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr23_+_45845423 | 1.33 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr18_-_26715156 | 1.33 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr3_+_32410746 | 1.32 |
ENSDART00000025496
|
rras
|
RAS related |
| chr23_+_38957738 | 1.31 |
ENSDART00000193480
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr21_-_22117085 | 1.29 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr1_-_27014872 | 1.28 |
ENSDART00000147414
ENSDART00000134032 ENSDART00000192087 ENSDART00000189111 ENSDART00000187348 ENSDART00000187248 |
cntln
|
centlein, centrosomal protein |
| chr21_+_21012210 | 1.28 |
ENSDART00000190317
|
ndr1
|
nodal-related 1 |
| chr9_-_8314028 | 1.27 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr21_+_39197628 | 1.23 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr16_-_24642814 | 1.23 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr21_-_35325466 | 1.21 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr2_-_40890004 | 1.20 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr13_-_4018888 | 1.19 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr2_-_57918314 | 1.19 |
ENSDART00000138265
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr6_+_21536131 | 1.18 |
ENSDART00000113911
ENSDART00000188472 |
micall1a
|
MICAL-like 1a |
| chr24_-_10021341 | 1.16 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr9_-_14273652 | 1.15 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr5_+_30392148 | 1.15 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr11_-_34577034 | 1.15 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr3_+_26244353 | 1.15 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr4_-_4592287 | 1.14 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr4_-_14192254 | 1.14 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr4_-_12930086 | 1.13 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr13_+_33462232 | 1.12 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr8_+_45334255 | 1.10 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr22_+_2510828 | 1.08 |
ENSDART00000115348
ENSDART00000145611 |
zgc:173726
|
zgc:173726 |
| chr8_-_53166975 | 1.08 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr6_+_12527725 | 1.08 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr23_+_32029304 | 1.08 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr22_-_20812822 | 1.08 |
ENSDART00000193778
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr1_+_54124209 | 1.07 |
ENSDART00000187730
|
LO017722.1
|
|
| chr7_-_24520866 | 1.06 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr14_+_15155684 | 1.05 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr10_-_34916208 | 1.05 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr18_-_6855991 | 1.05 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr22_+_2511045 | 1.05 |
ENSDART00000106425
|
zgc:173726
|
zgc:173726 |
| chr12_+_33320504 | 1.03 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr13_-_31008275 | 1.03 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr15_-_28569786 | 1.02 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr23_+_45785563 | 1.00 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr21_+_18907102 | 1.00 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr13_+_15656042 | 0.99 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr5_+_37749503 | 0.99 |
ENSDART00000148586
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr8_-_19051906 | 0.99 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr18_+_36582815 | 0.98 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr18_-_3527686 | 0.98 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr17_+_24318753 | 0.98 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr3_+_18936009 | 0.98 |
ENSDART00000140330
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr20_-_40367493 | 0.97 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr10_-_45210184 | 0.97 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr11_+_18175893 | 0.97 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr20_+_4221978 | 0.97 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr20_+_6535176 | 0.96 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr2_-_40890264 | 0.96 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr19_-_19379084 | 0.95 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr10_-_26202766 | 0.94 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr8_+_28358161 | 0.94 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr8_-_53044300 | 0.94 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr18_-_7677208 | 0.93 |
ENSDART00000092456
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr13_+_1131748 | 0.92 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr2_-_38992304 | 0.92 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr1_+_53321878 | 0.92 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_+_29548630 | 0.92 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr12_+_28955766 | 0.91 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr21_-_14692119 | 0.91 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr1_-_18811517 | 0.91 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr10_+_14963898 | 0.91 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr3_-_26244256 | 0.89 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr5_+_18014931 | 0.88 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_+_15890984 | 0.88 |
ENSDART00000158433
|
pank4
|
pantothenate kinase 4 |
| chr19_-_5865766 | 0.88 |
ENSDART00000191007
|
LO018585.1
|
|
| chr21_+_3796196 | 0.88 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr16_-_5873866 | 0.88 |
ENSDART00000136936
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr2_-_37632896 | 0.87 |
ENSDART00000008302
|
insra
|
insulin receptor a |
| chr15_+_21262917 | 0.86 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr9_-_3934963 | 0.86 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr7_-_735118 | 0.86 |
ENSDART00000126424
|
MAP3K11
|
si:cabz01078036.1 |
| chr25_+_34247107 | 0.85 |
ENSDART00000148507
|
bnip2
|
BCL2 interacting protein 2 |
| chr23_+_32028574 | 0.85 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr12_+_1609563 | 0.84 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr23_-_22072716 | 0.84 |
ENSDART00000054362
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr18_-_15532016 | 0.83 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr8_+_21437908 | 0.82 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr17_+_23730089 | 0.80 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr15_-_20839763 | 0.80 |
ENSDART00000141746
ENSDART00000182369 |
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr8_-_22538588 | 0.79 |
ENSDART00000144041
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr4_-_1720648 | 0.79 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr25_-_17587785 | 0.79 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr9_+_38457806 | 0.78 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr4_+_9911534 | 0.77 |
ENSDART00000109452
|
sbf1
|
SET binding factor 1 |
| chr8_+_42941555 | 0.77 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr5_+_31036911 | 0.77 |
ENSDART00000141463
ENSDART00000162121 |
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr8_+_50534948 | 0.77 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr8_+_7097929 | 0.77 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr7_+_9326234 | 0.77 |
ENSDART00000104536
|
chsy1
|
chondroitin sulfate synthase 1 |
| chr8_+_2543525 | 0.76 |
ENSDART00000129569
ENSDART00000128227 |
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr22_+_30137374 | 0.76 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr4_+_9911043 | 0.76 |
ENSDART00000159762
ENSDART00000091599 |
sbf1
|
SET binding factor 1 |
| chr21_-_36453594 | 0.74 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr17_+_23729717 | 0.74 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr14_-_45967981 | 0.74 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr24_+_24285751 | 0.74 |
ENSDART00000122294
|
ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr21_+_6394929 | 0.73 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr19_-_46037835 | 0.73 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr18_+_45573251 | 0.73 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr6_+_59642695 | 0.73 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr15_+_2856696 | 0.72 |
ENSDART00000163434
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr17_+_25197180 | 0.71 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr2_-_19321927 | 0.71 |
ENSDART00000182808
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr2_-_50126337 | 0.71 |
ENSDART00000009347
|
exoc3
|
exocyst complex component 3 |
| chr15_-_30857350 | 0.71 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr15_+_5132439 | 0.70 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr22_+_2751887 | 0.70 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr24_-_40901410 | 0.70 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr22_-_9906790 | 0.70 |
ENSDART00000105936
|
si:dkey-253d23.8
|
si:dkey-253d23.8 |
| chr11_+_18183220 | 0.70 |
ENSDART00000113468
|
LO018315.10
|
|
| chr23_+_38171186 | 0.70 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr6_+_40467562 | 0.69 |
ENSDART00000103868
|
zgc:152986
|
zgc:152986 |
| chr24_+_5789790 | 0.68 |
ENSDART00000189600
|
BX470065.1
|
|
| chr11_-_20988238 | 0.68 |
ENSDART00000155238
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr22_-_22147375 | 0.68 |
ENSDART00000149304
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr25_+_35132090 | 0.68 |
ENSDART00000154377
|
si:dkey-261m9.6
|
si:dkey-261m9.6 |
| chr17_-_1703259 | 0.67 |
ENSDART00000156489
|
xgb
|
x globin |
| chr7_-_9873652 | 0.67 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr25_+_34247353 | 0.67 |
ENSDART00000148914
|
bnip2
|
BCL2 interacting protein 2 |
| chr5_-_32396929 | 0.67 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr21_+_40695345 | 0.66 |
ENSDART00000143594
|
ccdc82
|
coiled-coil domain containing 82 |
| chr8_+_21297217 | 0.66 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr18_+_35229115 | 0.65 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr24_-_36187094 | 0.65 |
ENSDART00000111891
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr23_+_22597624 | 0.65 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr15_-_36727462 | 0.65 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr6_+_51713076 | 0.64 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr19_-_4785734 | 0.64 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_+_31324335 | 0.64 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_+_25650917 | 0.64 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr7_-_48391571 | 0.64 |
ENSDART00000180034
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr13_-_45811137 | 0.63 |
ENSDART00000190512
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr3_-_3439150 | 0.63 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr4_-_20081621 | 0.63 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr12_-_10508952 | 0.63 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr17_+_16429826 | 0.62 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr2_+_42005475 | 0.62 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr16_+_47207691 | 0.62 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr3_-_25268751 | 0.62 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr24_+_5789582 | 0.62 |
ENSDART00000141504
|
BX470065.1
|
|
| chr9_-_25425381 | 0.62 |
ENSDART00000129522
|
acvr2aa
|
activin A receptor type 2Aa |
| chr2_+_45511459 | 0.62 |
ENSDART00000132101
|
aknad1
|
AKNA domain containing 1 |
| chr10_+_44146287 | 0.61 |
ENSDART00000162176
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr4_-_72287764 | 0.61 |
ENSDART00000125452
ENSDART00000189437 |
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr22_+_1751640 | 0.61 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr8_-_39884359 | 0.60 |
ENSDART00000131372
|
mlec
|
malectin |
| chr6_+_2174082 | 0.60 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr16_+_13965923 | 0.60 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr4_-_837768 | 0.60 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr15_-_16076399 | 0.60 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr22_-_24757785 | 0.59 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr18_+_35128685 | 0.59 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr5_-_19444930 | 0.58 |
ENSDART00000136259
ENSDART00000188499 |
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr8_+_2656681 | 0.58 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr8_+_45381298 | 0.58 |
ENSDART00000149451
ENSDART00000110364 |
tti2
|
TELO2 interacting protein 2 |
| chr11_-_3494964 | 0.58 |
ENSDART00000162369
|
acap3b
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3b |
| chr18_+_8937718 | 0.57 |
ENSDART00000138115
|
si:dkey-5i3.1
|
si:dkey-5i3.1 |
| chr21_+_3897680 | 0.57 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr21_-_28737320 | 0.57 |
ENSDART00000098696
ENSDART00000124826 ENSDART00000125652 |
nrg2a
|
neuregulin 2a |
| chr20_+_48129759 | 0.57 |
ENSDART00000148494
|
tram2
|
translocation associated membrane protein 2 |
| chr5_-_54712159 | 0.57 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr21_-_14830353 | 0.57 |
ENSDART00000134687
|
pus1
|
pseudouridylate synthase 1 |
| chr22_-_9839003 | 0.57 |
ENSDART00000193573
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr20_-_40360571 | 0.57 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr4_-_48634516 | 0.57 |
ENSDART00000129731
|
znf1063
|
zinc finger protein 1063 |
| chr16_+_54209504 | 0.57 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr21_+_45502773 | 0.56 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr19_+_31532043 | 0.56 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr11_+_31323746 | 0.56 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx2_hmx3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.7 | 2.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 1.7 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 1.6 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.3 | 0.8 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.3 | 2.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.2 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.2 | 0.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 1.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 1.6 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.2 | 0.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 1.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 2.0 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 0.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 0.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.8 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 2.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.9 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.9 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.8 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.1 | 0.5 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 1.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.5 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 2.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 1.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.4 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.6 | GO:0002208 | somatic recombination of immunoglobulin genes involved in immune response(GO:0002204) somatic diversification of immunoglobulins involved in immune response(GO:0002208) immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.1 | 0.4 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.6 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 1.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.5 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 3.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.3 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 0.7 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.3 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.5 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.8 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0048798 | protein geranylgeranylation(GO:0018344) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 1.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.7 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.2 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.2 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 1.1 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.3 | GO:0018202 | histone H3-K36 methylation(GO:0010452) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.3 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 1.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 5.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.8 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.7 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 1.0 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.9 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.1 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0006826 | iron ion transport(GO:0006826) erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.8 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 2.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0032200 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.4 | 2.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 1.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 0.8 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 1.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.8 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 2.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 3.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 2.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.4 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 1.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 1.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 3.8 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 1.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.3 | 1.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 3.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 2.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.0 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.2 | 1.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.5 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 2.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.5 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 0.9 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 0.5 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.4 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.5 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.3 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.4 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.2 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 1.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 2.1 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 3.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 4.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |