Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
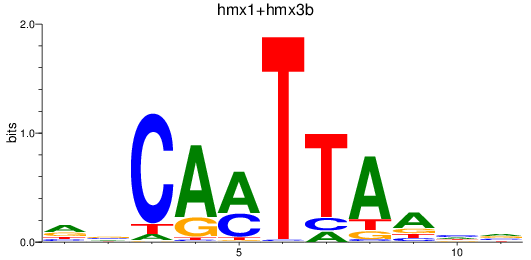
Results for hmx1+hmx3b
Z-value: 0.74
Transcription factors associated with hmx1+hmx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx1
|
ENSDARG00000095651 | H6 family homeobox 1 |
|
hmx3b
|
ENSDARG00000096716 | H6 family homeobox 3b |
|
hmx1
|
ENSDARG00000109725 | H6 family homeobox 1 |
|
hmx1
|
ENSDARG00000111765 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx1 | dr11_v1_chr1_-_40914752_40914752 | 0.91 | 6.5e-04 | Click! |
| hmx3b | dr11_v1_chr12_-_49154934_49154934 | 0.64 | 6.2e-02 | Click! |
Activity profile of hmx1+hmx3b motif
Sorted Z-values of hmx1+hmx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_26657404 | 1.67 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr11_+_30244356 | 1.51 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr14_-_3381303 | 1.44 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr11_-_7261717 | 1.29 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr3_+_60721342 | 1.28 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr12_+_2446837 | 1.27 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr5_-_42272517 | 1.14 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr20_+_28861435 | 1.06 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr8_-_23194408 | 1.05 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr18_+_9171778 | 1.04 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr25_+_18965430 | 1.01 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr7_+_71683853 | 0.99 |
ENSDART00000163002
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr18_-_5595546 | 0.96 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr15_-_1822548 | 0.91 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr7_+_32722227 | 0.90 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr20_+_28861629 | 0.80 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr14_+_14568437 | 0.78 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr1_+_34696503 | 0.77 |
ENSDART00000186106
|
CR339054.2
|
|
| chr1_+_52929185 | 0.75 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_+_24342303 | 0.67 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr5_+_11290851 | 0.67 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr13_-_50108337 | 0.65 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr24_+_38301080 | 0.63 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr5_-_14326959 | 0.62 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr23_-_18913032 | 0.60 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr15_-_14884332 | 0.58 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr22_-_6988102 | 0.58 |
ENSDART00000185618
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr14_-_33454595 | 0.58 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr9_-_31108285 | 0.54 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr25_-_10503043 | 0.54 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr6_-_37422841 | 0.54 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chrM_+_8130 | 0.53 |
ENSDART00000093609
|
mt-co2
|
cytochrome c oxidase II, mitochondrial |
| chr22_-_4649238 | 0.53 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr7_+_66565930 | 0.51 |
ENSDART00000154597
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr21_+_21906671 | 0.50 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr16_-_6391275 | 0.50 |
ENSDART00000060550
|
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr10_-_39153959 | 0.47 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr8_-_13013123 | 0.47 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr10_-_39154594 | 0.45 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr13_+_30874153 | 0.41 |
ENSDART00000112380
ENSDART00000189016 ENSDART00000134809 |
ercc6
|
excision repair cross-complementation group 6 |
| chr10_+_35439952 | 0.41 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr18_+_11970987 | 0.40 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr13_-_37109987 | 0.40 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_-_19649617 | 0.39 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr15_+_1134870 | 0.39 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr21_-_37973819 | 0.38 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr23_+_9353552 | 0.38 |
ENSDART00000163298
|
BX511246.1
|
|
| chr14_-_7137808 | 0.36 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr20_+_37295006 | 0.33 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr1_-_22512063 | 0.33 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr5_+_37032038 | 0.31 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr13_+_23176330 | 0.31 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr4_-_18741942 | 0.28 |
ENSDART00000145747
ENSDART00000066980 ENSDART00000186518 |
atxn10
|
ataxin 10 |
| chr10_+_16069987 | 0.27 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr24_-_27400017 | 0.26 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr6_-_6448519 | 0.26 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr20_-_34164278 | 0.25 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr4_+_14727018 | 0.25 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr6_-_40697585 | 0.24 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr25_+_3327071 | 0.24 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr25_+_3326885 | 0.23 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr4_-_76154252 | 0.19 |
ENSDART00000161269
|
BX957252.1
|
|
| chr4_+_13931733 | 0.18 |
ENSDART00000141742
ENSDART00000067175 |
pphln1
|
periphilin 1 |
| chr9_+_43799829 | 0.18 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr5_+_32835219 | 0.18 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr8_+_22472584 | 0.17 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr2_-_16380283 | 0.15 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr4_+_13931578 | 0.15 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr19_+_43604256 | 0.15 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr10_+_43039947 | 0.15 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr4_-_72609735 | 0.14 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr4_-_43388943 | 0.14 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr17_+_6563307 | 0.13 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr8_+_23194212 | 0.12 |
ENSDART00000028946
ENSDART00000186842 ENSDART00000137815 ENSDART00000136914 ENSDART00000144855 ENSDART00000182689 |
tpd52l2a
|
tumor protein D52-like 2a |
| chr22_+_1911269 | 0.11 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr18_+_44080948 | 0.10 |
ENSDART00000191036
|
BX908388.2
|
|
| chr18_-_25568994 | 0.09 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr13_+_30169681 | 0.09 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr14_-_9128919 | 0.07 |
ENSDART00000108641
|
sh2d1ab
|
SH2 domain containing 1A duplicate b |
| chr11_-_37691449 | 0.07 |
ENSDART00000185340
|
zgc:158258
|
zgc:158258 |
| chr19_-_6239248 | 0.07 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr21_+_21702623 | 0.06 |
ENSDART00000136840
|
or125-2
|
odorant receptor, family E, subfamily 125, member 2 |
| chr4_+_20576857 | 0.06 |
ENSDART00000125340
|
BX248410.2
|
|
| chr1_-_23596391 | 0.05 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr3_+_22327738 | 0.05 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr21_-_14803366 | 0.05 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr21_-_14966718 | 0.05 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr6_-_40352215 | 0.05 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr8_-_46486009 | 0.04 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr2_+_16652238 | 0.03 |
ENSDART00000091351
|
gk5
|
glycerol kinase 5 (putative) |
| chr22_+_724639 | 0.03 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr1_-_56032619 | 0.02 |
ENSDART00000143793
|
c3a.4
|
complement component c3a, duplicate 4 |
| chr10_+_40690411 | 0.00 |
ENSDART00000146865
|
taar19m
|
trace amine associated receptor 19m |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx1+hmx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.0 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.2 | 0.5 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 0.5 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.6 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.5 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 1.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.0 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.0 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.0 | 0.0 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |