Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
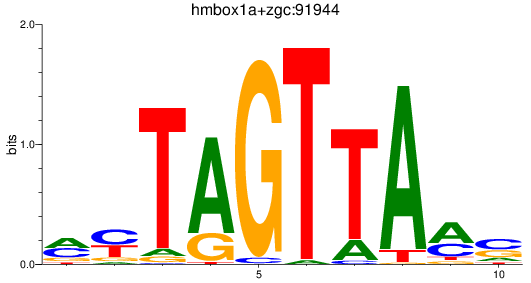
Results for hmbox1a+zgc:91944
Z-value: 1.08
Transcription factors associated with hmbox1a+zgc:91944
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmbox1a
|
ENSDARG00000027082 | homeobox containing 1a |
|
zgc
|
ENSDARG00000035887 | 91944 |
|
hmbox1a
|
ENSDARG00000109287 | homeobox containing 1 b |
|
zgc
|
ENSDARG00000114642 | 91944 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmbox1a | dr11_v1_chr17_-_16324565_16324565 | 0.90 | 1.0e-03 | Click! |
| CABZ01078261.1 | dr11_v1_chr20_+_49787584_49787584 | 0.80 | 9.7e-03 | Click! |
| zgc:91944 | dr11_v1_chr19_-_32600823_32600868 | -0.66 | 5.5e-02 | Click! |
Activity profile of hmbox1a+zgc:91944 motif
Sorted Z-values of hmbox1a+zgc:91944 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_30074332 | 2.14 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr15_-_1036878 | 1.79 |
ENSDART00000123844
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr5_-_15283509 | 1.62 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr18_+_35130416 | 1.56 |
ENSDART00000151595
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr10_-_26196383 | 1.47 |
ENSDART00000192925
|
fhdc3
|
FH2 domain containing 3 |
| chr24_-_9979342 | 1.47 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr9_+_22657221 | 1.44 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr24_+_18714212 | 1.40 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr9_+_29431763 | 1.37 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr10_-_26202766 | 1.35 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr15_+_6459847 | 1.31 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr19_+_31585917 | 1.29 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr5_+_31791001 | 1.29 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr2_-_21438492 | 1.25 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr5_-_67799821 | 1.21 |
ENSDART00000017881
|
eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr13_-_21672131 | 1.20 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr5_-_67799617 | 1.17 |
ENSDART00000192117
|
eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr13_+_39182099 | 1.16 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr5_-_34964830 | 1.16 |
ENSDART00000133170
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr5_-_11809404 | 1.14 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr7_+_35191220 | 1.12 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr1_+_30723380 | 1.11 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr23_-_14766902 | 1.10 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr12_-_33359052 | 1.10 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_+_23677179 | 1.09 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr2_+_29996650 | 1.08 |
ENSDART00000138050
ENSDART00000141026 |
rbm33b
|
RNA binding motif protein 33b |
| chr16_-_42965192 | 1.08 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr5_+_20257225 | 1.07 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr2_+_10160484 | 1.07 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr13_-_44852324 | 1.04 |
ENSDART00000136455
|
si:dkeyp-2e4.2
|
si:dkeyp-2e4.2 |
| chr19_+_31585341 | 1.03 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr4_-_5077158 | 1.02 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr1_+_44395976 | 1.01 |
ENSDART00000159686
ENSDART00000189905 ENSDART00000025145 |
unc93b1
|
unc-93 homolog B1, TLR signaling regulator |
| chr2_+_212059 | 0.99 |
ENSDART00000113021
|
dhx30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr2_-_27619954 | 0.99 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr18_-_20458840 | 0.99 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr16_+_45930962 | 0.99 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr9_-_41024282 | 0.98 |
ENSDART00000188260
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr8_-_22274509 | 0.97 |
ENSDART00000181805
ENSDART00000100063 |
nphp4
|
nephronophthisis 4 |
| chr24_-_11076400 | 0.96 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr9_-_49805218 | 0.95 |
ENSDART00000179290
|
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr19_+_26640096 | 0.94 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr17_-_11368662 | 0.94 |
ENSDART00000159061
ENSDART00000188694 ENSDART00000190932 |
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr20_-_39789846 | 0.94 |
ENSDART00000188418
|
rnf217
|
ring finger protein 217 |
| chr8_-_22274222 | 0.94 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr13_-_24429066 | 0.93 |
ENSDART00000147514
ENSDART00000057588 |
cep68
|
centrosomal protein 68 |
| chr8_-_28259766 | 0.92 |
ENSDART00000134226
ENSDART00000087037 ENSDART00000136973 |
ddx20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr6_+_44197348 | 0.92 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr12_-_13650344 | 0.92 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr7_+_36467796 | 0.91 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr3_-_47128096 | 0.90 |
ENSDART00000063287
|
qtrt1
|
queuine tRNA-ribosyltransferase 1 |
| chr7_-_16194952 | 0.88 |
ENSDART00000173739
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr17_+_28624321 | 0.86 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr19_-_20430892 | 0.85 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr23_-_36857964 | 0.84 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr4_+_26056548 | 0.83 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr4_-_14486822 | 0.83 |
ENSDART00000048821
|
plxnb2a
|
plexin b2a |
| chr14_+_31498439 | 0.80 |
ENSDART00000173281
|
phf6
|
PHD finger protein 6 |
| chr17_+_21902058 | 0.80 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr13_+_9612395 | 0.80 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr16_-_17699111 | 0.79 |
ENSDART00000108581
|
si:dkey-17m8.1
|
si:dkey-17m8.1 |
| chr3_+_18840810 | 0.78 |
ENSDART00000181137
ENSDART00000128626 ENSDART00000133332 |
tmem104
|
transmembrane protein 104 |
| chr11_+_25157374 | 0.77 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr18_-_341489 | 0.77 |
ENSDART00000168333
|
si:ch1073-83b15.1
|
si:ch1073-83b15.1 |
| chr21_-_36453417 | 0.73 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr17_-_15188440 | 0.73 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr16_+_33992418 | 0.72 |
ENSDART00000101885
ENSDART00000130540 |
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr7_-_16195144 | 0.72 |
ENSDART00000173492
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr22_-_16755885 | 0.72 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr7_+_13609457 | 0.72 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr13_+_8693410 | 0.70 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr2_-_2957970 | 0.69 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr13_-_24906307 | 0.68 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr4_-_5366566 | 0.68 |
ENSDART00000150788
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr19_+_43669122 | 0.68 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr13_+_46718518 | 0.66 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr13_+_40635844 | 0.66 |
ENSDART00000137310
|
hpse2
|
heparanase 2 |
| chr3_+_22335030 | 0.66 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr10_-_22912255 | 0.65 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr17_-_40876680 | 0.64 |
ENSDART00000127200
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr25_+_25737386 | 0.63 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr17_+_25197180 | 0.63 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr2_-_37743834 | 0.62 |
ENSDART00000088040
ENSDART00000191057 |
myo9b
|
myosin IXb |
| chr11_+_11030908 | 0.62 |
ENSDART00000111091
|
pla2r1
|
phospholipase A2 receptor 1 |
| chr7_+_52753067 | 0.62 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr9_+_29643036 | 0.62 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr16_+_29690708 | 0.62 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr1_-_23110740 | 0.62 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr7_-_25133783 | 0.61 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr21_+_45502621 | 0.61 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr8_-_22273819 | 0.61 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr21_-_2042037 | 0.60 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr13_+_28785814 | 0.60 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr13_-_31938512 | 0.60 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr7_-_38570878 | 0.59 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr3_-_11878490 | 0.59 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr2_-_21621878 | 0.59 |
ENSDART00000189540
|
zgc:55781
|
zgc:55781 |
| chr10_+_41939963 | 0.58 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr16_-_43011470 | 0.58 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr17_+_30591287 | 0.58 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr1_-_55238610 | 0.57 |
ENSDART00000110818
|
FO704915.1
|
|
| chr10_+_45302425 | 0.57 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr18_-_30021479 | 0.55 |
ENSDART00000144562
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr6_-_31325400 | 0.55 |
ENSDART00000188869
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr9_-_54248182 | 0.54 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr9_-_1483879 | 0.53 |
ENSDART00000093415
|
rbm45
|
RNA binding motif protein 45 |
| chr13_+_8840772 | 0.53 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr2_-_24061575 | 0.53 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr19_+_32201991 | 0.52 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr24_-_20321012 | 0.52 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr2_-_43123949 | 0.51 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr8_+_10339869 | 0.51 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr12_+_12789853 | 0.50 |
ENSDART00000064839
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr19_-_26923957 | 0.50 |
ENSDART00000182390
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr18_-_2727764 | 0.50 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr11_-_40504170 | 0.50 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr21_+_45502773 | 0.49 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr3_-_30186296 | 0.49 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr8_+_47100863 | 0.49 |
ENSDART00000114811
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr12_-_18577774 | 0.49 |
ENSDART00000078166
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr4_+_72798545 | 0.49 |
ENSDART00000181727
|
MYRFL
|
myelin regulatory factor-like |
| chr10_-_28513861 | 0.49 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr5_+_19448078 | 0.48 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr23_-_17101360 | 0.48 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr7_-_62003831 | 0.47 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr10_-_2524917 | 0.47 |
ENSDART00000188642
|
CU856539.1
|
|
| chr14_+_16151636 | 0.47 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr24_-_7587401 | 0.47 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_+_23701613 | 0.46 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr12_-_20665164 | 0.46 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr12_+_26538861 | 0.46 |
ENSDART00000152955
|
si:dkey-57h18.1
|
si:dkey-57h18.1 |
| chr12_-_26538823 | 0.46 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr17_+_4325693 | 0.46 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr17_-_14451718 | 0.46 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr17_-_7818944 | 0.46 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr7_+_53199763 | 0.46 |
ENSDART00000160097
|
cdh28
|
cadherin 28 |
| chr20_+_32552912 | 0.45 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr5_+_36439405 | 0.45 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr11_-_36230146 | 0.45 |
ENSDART00000135888
ENSDART00000189782 |
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr23_+_13528053 | 0.45 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr3_-_26806032 | 0.44 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr25_+_25085349 | 0.43 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr2_-_3403020 | 0.43 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr12_+_19281189 | 0.43 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr8_+_20140321 | 0.43 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr12_-_27588299 | 0.43 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr25_+_16356083 | 0.42 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr8_+_3530761 | 0.42 |
ENSDART00000081272
|
gcn1
|
GCN1 eIF2 alpha kinase activator homolog |
| chr18_-_44331041 | 0.42 |
ENSDART00000168619
|
prdm10
|
PR domain containing 10 |
| chr16_-_1757521 | 0.42 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr22_+_18315490 | 0.41 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr1_-_29061285 | 0.41 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr18_-_18584839 | 0.41 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr21_+_33459524 | 0.41 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr18_-_31051847 | 0.40 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr20_-_23494695 | 0.40 |
ENSDART00000058524
ENSDART00000146203 |
anapc4
|
anaphase promoting complex subunit 4 |
| chr12_+_35011899 | 0.40 |
ENSDART00000153007
ENSDART00000153020 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr7_-_23848012 | 0.40 |
ENSDART00000146104
ENSDART00000175108 |
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr23_-_44786844 | 0.39 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr9_-_55772937 | 0.39 |
ENSDART00000159192
|
akap17a
|
A kinase (PRKA) anchor protein 17A |
| chr20_-_47051996 | 0.39 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr4_+_9011448 | 0.37 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr18_+_13077800 | 0.37 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr16_+_33121106 | 0.37 |
ENSDART00000110195
|
akirin1
|
akirin 1 |
| chr4_-_9664995 | 0.37 |
ENSDART00000180105
ENSDART00000029084 |
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr19_-_3874986 | 0.37 |
ENSDART00000161830
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr13_-_226109 | 0.36 |
ENSDART00000161705
ENSDART00000172744 ENSDART00000163902 ENSDART00000158208 |
rtn4b
|
reticulon 4b |
| chr23_-_30960506 | 0.36 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr3_-_44059902 | 0.35 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr24_-_35269991 | 0.35 |
ENSDART00000185424
|
sntg1
|
syntrophin, gamma 1 |
| chr24_-_18809433 | 0.35 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr23_-_18982499 | 0.35 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr5_-_29559758 | 0.34 |
ENSDART00000051471
ENSDART00000167115 |
nelfb
|
negative elongation factor complex member B |
| chr2_+_30960351 | 0.34 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr21_-_26028205 | 0.34 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr13_+_48359573 | 0.33 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr19_+_7559901 | 0.33 |
ENSDART00000141189
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr19_-_20099394 | 0.33 |
ENSDART00000126173
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr2_+_33926606 | 0.33 |
ENSDART00000111430
|
kif2c
|
kinesin family member 2C |
| chr23_+_25868637 | 0.33 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr7_+_17938128 | 0.32 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr3_-_23512285 | 0.32 |
ENSDART00000159151
|
BX682558.1
|
|
| chr3_-_36246899 | 0.32 |
ENSDART00000141106
ENSDART00000059506 ENSDART00000158701 |
gps1
|
G protein pathway suppressor 1 |
| chr9_-_25328527 | 0.31 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr17_-_25382367 | 0.31 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr17_-_37184655 | 0.31 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr19_+_12406583 | 0.31 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr1_+_513986 | 0.30 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr15_+_17074185 | 0.29 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr17_+_5985933 | 0.28 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr6_-_48473694 | 0.28 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr20_-_1378514 | 0.28 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr5_+_25733774 | 0.28 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr21_+_37090585 | 0.28 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr6_+_37441789 | 0.28 |
ENSDART00000157317
|
gtpbp6
|
GTP binding protein 6 (putative) |
| chr14_-_36437249 | 0.28 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr2_-_22631498 | 0.27 |
ENSDART00000146425
|
stk25b
|
serine/threonine kinase 25b |
| chr12_+_40945319 | 0.27 |
ENSDART00000183435
|
CDH18
|
cadherin 18 |
| chr18_-_502722 | 0.26 |
ENSDART00000185757
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr15_+_19335646 | 0.26 |
ENSDART00000062569
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmbox1a+zgc:91944
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 1.0 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 2.0 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 0.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 1.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.6 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.4 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.7 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 1.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.3 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 2.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 1.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.4 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.4 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.4 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 2.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.6 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.9 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.2 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 1.1 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.7 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.1 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.1 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 1.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.6 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 2.0 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 1.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.5 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 2.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.0 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.3 | GO:0032356 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.5 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.2 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 3.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |