Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
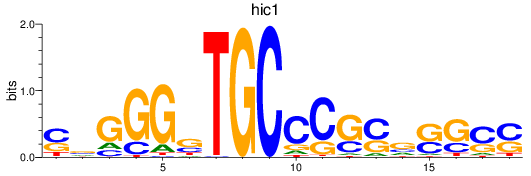
Results for hic1
Z-value: 1.02
Transcription factors associated with hic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1
|
ENSDARG00000055493 | hypermethylated in cancer 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic1 | dr11_v1_chr15_+_25683069_25683069 | 0.86 | 3.2e-03 | Click! |
Activity profile of hic1 motif
Sorted Z-values of hic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77561679 | 2.36 |
ENSDART00000180809
|
AL935186.9
|
|
| chr4_+_55778679 | 2.26 |
ENSDART00000183009
|
CT583728.19
|
|
| chr4_+_55794876 | 2.26 |
ENSDART00000189043
|
CT583728.17
|
|
| chr4_+_55810436 | 2.26 |
ENSDART00000182875
|
CT583728.16
|
|
| chr4_-_68568233 | 2.26 |
ENSDART00000184284
|
BX548011.1
|
|
| chr4_-_55728559 | 2.16 |
ENSDART00000186201
|
CT583728.14
|
|
| chr23_+_45845159 | 1.42 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr23_+_45845423 | 1.11 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr16_+_25285998 | 1.09 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr1_-_8928841 | 1.01 |
ENSDART00000103652
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr5_+_393738 | 1.00 |
ENSDART00000161456
|
june
|
JunE proto-oncogene, AP-1 transcription factor subunit |
| chr1_+_45839927 | 0.97 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr6_+_28203 | 0.96 |
ENSDART00000191561
|
CZQB01141835.1
|
|
| chr17_-_8976307 | 0.96 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr16_+_41067586 | 0.87 |
ENSDART00000181876
|
scap
|
SREBF chaperone |
| chr23_-_31060350 | 0.84 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr8_+_54137350 | 0.68 |
ENSDART00000164153
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr3_+_17653784 | 0.68 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr2_-_19354622 | 0.64 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr3_+_37083765 | 0.59 |
ENSDART00000125611
|
retreg3
|
reticulophagy regulator family member 3 |
| chr1_+_18863060 | 0.59 |
ENSDART00000139241
|
rnf38
|
ring finger protein 38 |
| chr9_-_14992730 | 0.52 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr15_-_9421481 | 0.51 |
ENSDART00000189045
ENSDART00000177158 |
sacs
|
sacsin molecular chaperone |
| chr11_-_6420917 | 0.48 |
ENSDART00000193717
|
FO681393.2
|
|
| chr5_-_42123794 | 0.46 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr17_-_49412313 | 0.39 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr19_-_38830582 | 0.38 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr10_-_2682198 | 0.36 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr2_-_36819624 | 0.35 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr8_-_13678415 | 0.32 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr2_+_21063660 | 0.32 |
ENSDART00000022765
|
riok1
|
RIO kinase 1 (yeast) |
| chr4_-_797831 | 0.29 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr25_-_1687481 | 0.28 |
ENSDART00000163553
ENSDART00000179460 |
mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr19_-_12967986 | 0.28 |
ENSDART00000151064
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr7_-_24995631 | 0.27 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr12_-_979789 | 0.23 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr13_+_421231 | 0.23 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr12_-_54375 | 0.21 |
ENSDART00000152304
|
si:ch1073-357b18.4
|
si:ch1073-357b18.4 |
| chr3_+_46762703 | 0.17 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr19_+_17642356 | 0.14 |
ENSDART00000176431
|
CR382334.1
|
|
| chr14_-_48588422 | 0.09 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr6_-_1874664 | 0.09 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr22_-_18116635 | 0.08 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr3_-_56896702 | 0.03 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr5_+_62723233 | 0.01 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr16_+_50006145 | 0.01 |
ENSDART00000049375
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.6 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.9 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.2 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.7 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 1.0 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.7 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0032934 | sterol binding(GO:0032934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |