Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
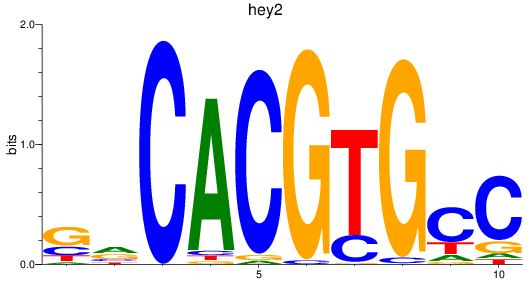
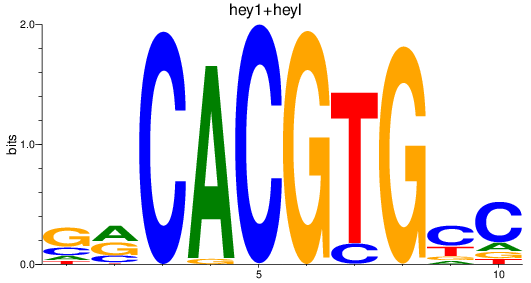
Results for hey2_hey1+heyl
Z-value: 0.73
Transcription factors associated with hey2_hey1+heyl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hey2
|
ENSDARG00000013441 | hes-related family bHLH transcription factor with YRPW motif 2 |
|
heyl
|
ENSDARG00000055798 | hes related family bHLH transcription factor with YRPW motif like |
|
hey1
|
ENSDARG00000070538 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
heyl
|
ENSDARG00000112770 | hes related family bHLH transcription factor with YRPW motif like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| heyl | dr11_v1_chr19_+_32553874_32553874 | 0.66 | 5.3e-02 | Click! |
| hey2 | dr11_v1_chr20_-_39596338_39596338 | -0.41 | 2.8e-01 | Click! |
| hey1 | dr11_v1_chr19_-_31802296_31802296 | 0.40 | 2.9e-01 | Click! |
Activity profile of hey2_hey1+heyl motif
Sorted Z-values of hey2_hey1+heyl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_7607114 | 1.20 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr19_-_24555935 | 1.18 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr4_-_13580348 | 1.10 |
ENSDART00000067160
|
opn1sw1
|
opsin 1 (cone pigments), short-wave-sensitive 1 |
| chr19_-_24555623 | 1.10 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr1_-_39943596 | 0.98 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr9_-_56272465 | 0.91 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_+_12968101 | 0.89 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr24_-_12938922 | 0.85 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr17_+_25414033 | 0.84 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr11_-_3552067 | 0.79 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr16_-_45069882 | 0.78 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr25_+_18964782 | 0.76 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr13_+_32148338 | 0.75 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr14_+_14836468 | 0.74 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr19_+_42470396 | 0.71 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr8_+_17184602 | 0.71 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr21_-_20765338 | 0.68 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr24_-_8732519 | 0.68 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr3_+_23488652 | 0.66 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr14_-_498979 | 0.64 |
ENSDART00000171976
|
spry1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr18_+_14693682 | 0.63 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr7_+_38750871 | 0.62 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr12_-_6159545 | 0.62 |
ENSDART00000152487
|
rltgr
|
RAMP-like triterpene glycoside receptor |
| chr16_+_23960744 | 0.59 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr16_+_23960933 | 0.57 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr7_-_30082931 | 0.56 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr20_-_48485354 | 0.56 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr23_+_36063599 | 0.55 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr14_+_46313135 | 0.53 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr22_+_16497670 | 0.53 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr20_+_45893173 | 0.52 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr6_-_60147517 | 0.52 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr22_-_12160283 | 0.51 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr18_-_21218851 | 0.51 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr14_+_24215046 | 0.51 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr10_-_7988396 | 0.50 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr23_-_21463788 | 0.49 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr7_+_49715750 | 0.49 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr20_+_34915945 | 0.48 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr7_-_73752955 | 0.48 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr21_-_42100471 | 0.47 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_+_42469058 | 0.47 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr6_-_24301324 | 0.46 |
ENSDART00000171401
|
BX927081.1
|
|
| chr16_+_20915319 | 0.46 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr21_+_6780340 | 0.45 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr14_+_35748385 | 0.44 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr6_+_42819337 | 0.44 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr4_+_8569199 | 0.44 |
ENSDART00000165181
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr2_+_25019387 | 0.43 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr8_-_18239494 | 0.43 |
ENSDART00000079989
ENSDART00000022959 |
guk1b
|
guanylate kinase 1b |
| chr11_+_3959495 | 0.43 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr15_+_1148074 | 0.42 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr18_+_17428506 | 0.41 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr20_-_35246150 | 0.41 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr18_-_17020231 | 0.40 |
ENSDART00000129146
|
tbc1d15
|
TBC1 domain family, member 15 |
| chr2_-_42375275 | 0.39 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr5_+_4366431 | 0.39 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr18_+_17428258 | 0.39 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr3_+_32526263 | 0.39 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr8_-_25247284 | 0.39 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr20_-_147574 | 0.38 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr9_-_22281854 | 0.38 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr7_-_40993456 | 0.37 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr1_-_22834824 | 0.37 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr23_-_29824146 | 0.36 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr14_-_32016615 | 0.36 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr3_+_28939759 | 0.36 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr16_+_7154266 | 0.35 |
ENSDART00000168830
|
bmper
|
BMP binding endothelial regulator |
| chr14_+_36223097 | 0.35 |
ENSDART00000186872
|
pitx2
|
paired-like homeodomain 2 |
| chr6_-_10591607 | 0.35 |
ENSDART00000151376
|
dnah9l
|
dynein, axonemal, heavy polypeptide 9 like |
| chr25_+_37268900 | 0.35 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr1_-_50859053 | 0.35 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr3_+_54887782 | 0.35 |
ENSDART00000018205
|
aanat2
|
arylalkylamine N-acetyltransferase 2 |
| chr6_+_42818963 | 0.34 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr19_-_791016 | 0.34 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr8_-_23081511 | 0.34 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr12_+_47663419 | 0.34 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr1_+_7956030 | 0.33 |
ENSDART00000159655
|
CR855320.1
|
|
| chr7_-_30174882 | 0.33 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr1_-_35916247 | 0.33 |
ENSDART00000181541
|
smad1
|
SMAD family member 1 |
| chr19_+_43119698 | 0.33 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr24_-_33756003 | 0.33 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr23_+_21459263 | 0.32 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr10_+_17026870 | 0.32 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr3_+_20156956 | 0.32 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr25_-_6223567 | 0.32 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr22_+_28446365 | 0.32 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr12_+_24342303 | 0.31 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr19_+_43563179 | 0.30 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr15_-_2652640 | 0.30 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr1_+_45783886 | 0.30 |
ENSDART00000132555
|
pkn1a
|
protein kinase N1a |
| chr14_+_35748206 | 0.29 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr1_+_17376922 | 0.29 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr16_+_20895904 | 0.29 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr10_+_158590 | 0.29 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr14_-_30387894 | 0.28 |
ENSDART00000176136
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr6_+_22597362 | 0.28 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr6_-_44711942 | 0.28 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr17_-_17948587 | 0.28 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr11_-_16975190 | 0.28 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr3_-_62380146 | 0.28 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr18_-_44129151 | 0.28 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr4_+_17417111 | 0.28 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr4_-_21652643 | 0.27 |
ENSDART00000066898
|
rps16
|
ribosomal protein S16 |
| chr13_+_24662238 | 0.27 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr18_-_39583601 | 0.27 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr4_-_211714 | 0.26 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr19_-_22346582 | 0.26 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr25_-_21092222 | 0.26 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr24_-_42090635 | 0.26 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr2_+_29976419 | 0.26 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr9_-_22232902 | 0.26 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr14_-_42231293 | 0.25 |
ENSDART00000185486
|
BX890543.1
|
|
| chr11_+_24900123 | 0.25 |
ENSDART00000044987
ENSDART00000148023 |
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr14_-_46675850 | 0.25 |
ENSDART00000113285
|
tapt1a
|
transmembrane anterior posterior transformation 1a |
| chr9_-_44295071 | 0.25 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr22_-_94352 | 0.24 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr9_-_22240052 | 0.24 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr9_+_41821613 | 0.24 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr19_+_31183495 | 0.24 |
ENSDART00000088618
|
meox2b
|
mesenchyme homeobox 2b |
| chr13_+_281214 | 0.24 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr22_-_17489040 | 0.24 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr20_+_32406011 | 0.23 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr6_-_47953829 | 0.23 |
ENSDART00000157264
|
si:dkey-166d12.2
|
si:dkey-166d12.2 |
| chr2_+_301898 | 0.23 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr11_+_40032790 | 0.23 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr10_-_11012000 | 0.23 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr12_+_30200652 | 0.23 |
ENSDART00000193405
ENSDART00000132335 |
ablim1b
|
actin binding LIM protein 1b |
| chr5_-_61349059 | 0.22 |
ENSDART00000136553
|
si:ch211-209a2.2
|
si:ch211-209a2.2 |
| chr21_-_30408775 | 0.22 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr20_-_2949028 | 0.22 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr13_-_18637244 | 0.21 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr4_+_36489448 | 0.21 |
ENSDART00000143181
|
znf1149
|
zinc finger protein 1149 |
| chr2_+_16160906 | 0.21 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr2_-_5505094 | 0.21 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr18_-_19103929 | 0.21 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr20_-_54259780 | 0.20 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr21_-_20711739 | 0.20 |
ENSDART00000190918
|
BX530077.1
|
|
| chr19_+_24882845 | 0.20 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr24_-_22702017 | 0.20 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr19_+_22850657 | 0.20 |
ENSDART00000130472
|
pkhd1l1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
| chr18_+_40993369 | 0.20 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr5_+_36932718 | 0.20 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr13_-_37519774 | 0.20 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr19_+_19750101 | 0.20 |
ENSDART00000168041
ENSDART00000170697 |
hoxa9a
CR382300.2
|
homeobox A9a |
| chr14_-_8795081 | 0.20 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr8_+_23213320 | 0.20 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr18_-_12052132 | 0.19 |
ENSDART00000074361
|
zgc:110789
|
zgc:110789 |
| chr3_+_1223824 | 0.19 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr24_+_2519761 | 0.19 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr19_-_35361556 | 0.19 |
ENSDART00000012167
|
ndufs5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5 |
| chr11_+_25638172 | 0.19 |
ENSDART00000114226
ENSDART00000143677 |
grm6b
|
glutamate receptor, metabotropic 6b |
| chr9_-_785444 | 0.19 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr7_+_54605640 | 0.19 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr6_-_38818582 | 0.19 |
ENSDART00000149833
|
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr25_+_3328487 | 0.19 |
ENSDART00000181143
|
ldhbb
|
lactate dehydrogenase Bb |
| chr19_+_6938289 | 0.19 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr4_-_31064584 | 0.18 |
ENSDART00000187247
ENSDART00000134357 ENSDART00000166459 |
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr12_+_30586599 | 0.18 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr20_-_25486384 | 0.18 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr24_-_17023392 | 0.18 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr5_+_22677786 | 0.18 |
ENSDART00000142112
|
vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr2_-_47904043 | 0.18 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr2_-_9489611 | 0.18 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr19_+_7152966 | 0.18 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr13_+_29771463 | 0.18 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr7_+_67467702 | 0.18 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr17_-_23674495 | 0.18 |
ENSDART00000122209
|
ptena
|
phosphatase and tensin homolog A |
| chr10_-_41450367 | 0.18 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr17_+_19499157 | 0.17 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr12_+_42574148 | 0.17 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr2_-_10188598 | 0.17 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr15_-_36533322 | 0.17 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr2_+_6181383 | 0.17 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr13_+_4205724 | 0.17 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr4_-_5913338 | 0.17 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr8_+_40628926 | 0.17 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr8_+_46536893 | 0.17 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr2_-_31614277 | 0.17 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr16_-_560574 | 0.17 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr12_-_7655322 | 0.16 |
ENSDART00000181162
|
ank3b
|
ankyrin 3b |
| chr1_-_54765262 | 0.16 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr24_-_38787457 | 0.16 |
ENSDART00000112318
ENSDART00000154461 |
ntn2
|
netrin 2 |
| chr4_+_5180650 | 0.16 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr1_+_54766943 | 0.16 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr4_-_5912951 | 0.16 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr1_-_41192059 | 0.16 |
ENSDART00000084665
ENSDART00000135369 |
dok7
|
docking protein 7 |
| chr25_+_37443194 | 0.16 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr18_+_40993196 | 0.16 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr20_-_27733683 | 0.16 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr9_+_32872690 | 0.16 |
ENSDART00000020798
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr10_-_6775271 | 0.16 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr4_-_44611273 | 0.16 |
ENSDART00000156793
|
znf1115
|
zinc finger protein 1115 |
| chr1_-_30457062 | 0.15 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr11_-_15090564 | 0.15 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr22_-_20105969 | 0.15 |
ENSDART00000088687
|
rxfp3.2b
|
relaxin/insulin-like family peptide receptor 3.2b |
| chr19_+_19756425 | 0.15 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr12_+_19384615 | 0.15 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr4_-_13921185 | 0.15 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr4_-_68913650 | 0.15 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hey2_hey1+heyl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 0.8 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.9 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.2 | 0.6 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 0.5 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 0.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.8 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.9 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 2.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.2 | GO:0021985 | neurohypophysis development(GO:0021985) |
| 0.1 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.7 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.7 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.4 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.8 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.0 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.1 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 2.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.5 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.2 | 0.9 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.8 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.5 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |