Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
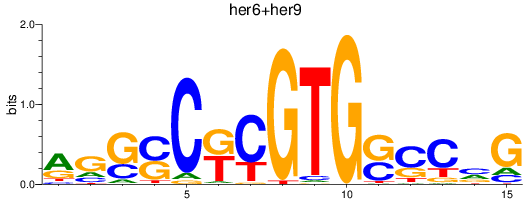
Results for her6+her9
Z-value: 0.59
Transcription factors associated with her6+her9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her6
|
ENSDARG00000006514 | hairy-related 6 |
|
her9
|
ENSDARG00000056438 | hairy-related 9 |
|
her6
|
ENSDARG00000111099 | hairy-related 6 |
|
her9
|
ENSDARG00000115775 | hairy-related 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her9 | dr11_v1_chr23_-_23401305_23401305 | -0.96 | 3.4e-05 | Click! |
| her6 | dr11_v1_chr6_-_36552844_36552844 | -0.86 | 2.9e-03 | Click! |
Activity profile of her6+her9 motif
Sorted Z-values of her6+her9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77551860 | 2.07 |
ENSDART00000188176
|
AL935186.6
|
|
| chr4_-_77563411 | 2.04 |
ENSDART00000186841
|
AL935186.8
|
|
| chr20_-_16498991 | 1.58 |
ENSDART00000104137
|
ches1
|
checkpoint suppressor 1 |
| chr12_+_48340133 | 1.48 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr1_+_49955869 | 0.94 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr16_+_48729947 | 0.94 |
ENSDART00000049051
|
brd2b
|
bromodomain containing 2b |
| chr7_-_6438544 | 0.91 |
ENSDART00000173180
|
zgc:165555
|
zgc:165555 |
| chr13_-_24826607 | 0.90 |
ENSDART00000087786
ENSDART00000186951 |
slka
|
STE20-like kinase a |
| chr14_-_32520295 | 0.90 |
ENSDART00000165463
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr7_+_24528430 | 0.85 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr9_+_500052 | 0.78 |
ENSDART00000166707
|
CU984600.1
|
|
| chr19_-_35450661 | 0.78 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr4_+_68562464 | 0.75 |
ENSDART00000192954
|
BX548011.4
|
|
| chr18_+_41772474 | 0.69 |
ENSDART00000097547
|
trpc6b
|
transient receptor potential cation channel, subfamily C, member 6b |
| chr8_+_36509885 | 0.67 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr18_+_3169579 | 0.61 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_-_2942900 | 0.60 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr5_-_12560569 | 0.59 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr21_+_1647990 | 0.54 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr5_-_29122834 | 0.54 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr22_-_193234 | 0.52 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr18_-_14274803 | 0.51 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr11_-_12800945 | 0.50 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr5_-_54395488 | 0.49 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr11_-_12801157 | 0.48 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr13_+_35925490 | 0.48 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr20_-_6196989 | 0.48 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr23_+_36616717 | 0.44 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr24_+_39537565 | 0.44 |
ENSDART00000140868
ENSDART00000145358 |
si:dkey-161j23.5
|
si:dkey-161j23.5 |
| chr5_+_1965296 | 0.39 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr25_+_27744293 | 0.38 |
ENSDART00000103519
ENSDART00000149456 |
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr8_+_12925385 | 0.38 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr1_-_54107321 | 0.37 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr5_-_29122615 | 0.35 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr3_+_14641962 | 0.32 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr3_-_11828206 | 0.30 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr5_-_36916790 | 0.30 |
ENSDART00000143827
|
kptn
|
kaptin (actin binding protein) |
| chr25_-_21280584 | 0.29 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr21_-_44731865 | 0.29 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr7_+_31374383 | 0.29 |
ENSDART00000085505
|
ctdspl2b
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2b |
| chr17_-_3986236 | 0.26 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr3_-_8765165 | 0.25 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr7_+_17938128 | 0.25 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr14_-_41075262 | 0.24 |
ENSDART00000180518
|
drp2
|
dystrophin related protein 2 |
| chr20_-_40766387 | 0.23 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr10_+_29138021 | 0.23 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr9_-_43538328 | 0.23 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr8_-_912821 | 0.15 |
ENSDART00000082296
|
ptger4a
|
prostaglandin E receptor 4 (subtype EP4) a |
| chr1_-_52461322 | 0.13 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr25_+_388258 | 0.10 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr23_-_6765653 | 0.10 |
ENSDART00000192310
|
FP102169.1
|
|
| chr9_+_30213144 | 0.10 |
ENSDART00000139920
|
senp7a
|
SUMO1/sentrin specific peptidase 7a |
| chr12_+_19362335 | 0.09 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr3_+_431208 | 0.07 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr13_-_45022527 | 0.07 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr12_+_36390422 | 0.07 |
ENSDART00000193826
|
galr2a
|
galanin receptor 2a |
| chr12_+_19362547 | 0.06 |
ENSDART00000180148
|
gspt1
|
G1 to S phase transition 1 |
| chr3_-_58823762 | 0.04 |
ENSDART00000101253
|
luc7l3
|
LUC7-like 3 pre-mRNA splicing factor |
| chr4_+_332709 | 0.04 |
ENSDART00000067486
|
tulp4a
|
tubby like protein 4a |
| chr22_+_19311411 | 0.03 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr25_+_19954576 | 0.01 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr14_+_12109535 | 0.01 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
Network of associatons between targets according to the STRING database.
First level regulatory network of her6+her9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 0.5 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.6 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.9 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 0.5 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |