Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
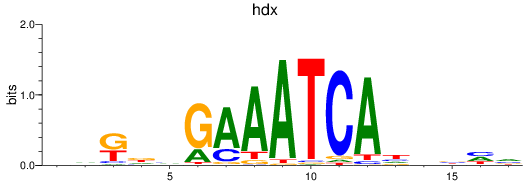
Results for hdx
Z-value: 1.21
Transcription factors associated with hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hdx
|
ENSDARG00000079382 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hdx | dr11_v1_chr14_-_6225336_6225336 | -0.05 | 8.9e-01 | Click! |
Activity profile of hdx motif
Sorted Z-values of hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_24382651 | 2.54 |
ENSDART00000123789
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr21_-_2322102 | 1.87 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr9_-_12888082 | 1.68 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr21_-_7940043 | 1.58 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr17_-_6618574 | 1.53 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr2_+_37295088 | 1.37 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr17_-_2596125 | 1.32 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr23_+_37579107 | 1.29 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr3_-_3413669 | 1.27 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr17_-_6641535 | 1.26 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr10_-_36793412 | 1.23 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr16_-_26820634 | 1.18 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_-_8928841 | 1.13 |
ENSDART00000103652
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr6_-_2133737 | 1.09 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr5_-_38342992 | 1.07 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr15_-_26887028 | 1.06 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr5_+_27267186 | 1.04 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr22_-_11054244 | 1.02 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr25_+_15997957 | 1.01 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr8_+_2773776 | 0.99 |
ENSDART00000156796
ENSDART00000154529 ENSDART00000155092 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr10_-_25570647 | 0.99 |
ENSDART00000134215
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr16_+_4181105 | 0.98 |
ENSDART00000180921
|
yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr14_+_34495216 | 0.96 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr7_+_13684012 | 0.94 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr2_+_34967210 | 0.93 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr2_+_45696743 | 0.91 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr12_+_30788912 | 0.90 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr23_-_478201 | 0.89 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr10_+_44373349 | 0.88 |
ENSDART00000172191
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr2_+_5300405 | 0.88 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr13_+_31550185 | 0.88 |
ENSDART00000127843
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr2_-_40890004 | 0.88 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr19_-_3488860 | 0.87 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr6_-_18751696 | 0.87 |
ENSDART00000171537
|
tnrc6c2
|
trinucleotide repeat containing 6C2 |
| chr22_-_23267893 | 0.85 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr23_+_19655301 | 0.84 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr15_+_42235449 | 0.84 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr24_-_27409599 | 0.83 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr14_-_30490763 | 0.82 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr13_-_35459928 | 0.81 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr6_+_54687495 | 0.80 |
ENSDART00000189514
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr16_+_19029297 | 0.79 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr21_+_39336285 | 0.79 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr10_-_7734939 | 0.79 |
ENSDART00000075563
|
snx24
|
sorting nexin 24 |
| chr3_-_27072179 | 0.78 |
ENSDART00000156556
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr5_-_64454459 | 0.78 |
ENSDART00000172321
ENSDART00000168030 |
brd3b
|
bromodomain containing 3b |
| chr6_+_21005725 | 0.78 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr5_+_30596632 | 0.77 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr9_+_8364553 | 0.76 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr23_-_42810664 | 0.74 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr13_+_39297802 | 0.73 |
ENSDART00000133636
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr5_-_3885727 | 0.73 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr7_+_38962459 | 0.73 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr2_-_40890264 | 0.72 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr13_+_15580758 | 0.72 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr8_-_39884359 | 0.72 |
ENSDART00000131372
|
mlec
|
malectin |
| chr5_+_61941610 | 0.71 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr25_-_36261836 | 0.71 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr10_-_18492617 | 0.71 |
ENSDART00000179936
ENSDART00000193799 ENSDART00000193198 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr16_-_35329803 | 0.71 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr17_+_6536152 | 0.70 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr17_-_6536466 | 0.69 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr5_+_63302660 | 0.68 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr3_+_16724614 | 0.68 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr8_-_4010887 | 0.68 |
ENSDART00000163678
|
mtmr3
|
myotubularin related protein 3 |
| chr21_+_17051478 | 0.67 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_+_26053044 | 0.67 |
ENSDART00000039877
|
SCYL2
|
si:ch211-244b2.1 |
| chr25_+_33072676 | 0.67 |
ENSDART00000182885
ENSDART00000181516 |
tln2b
|
talin 2b |
| chr13_-_24257631 | 0.66 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr8_-_53044300 | 0.66 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_+_25683069 | 0.66 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr16_+_3067134 | 0.66 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr7_+_20030888 | 0.65 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr14_+_30398546 | 0.65 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr22_-_5171362 | 0.65 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr10_-_32663759 | 0.65 |
ENSDART00000126727
|
atg101
|
autophagy related 101 |
| chr1_+_59314675 | 0.65 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr16_+_38820486 | 0.63 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr5_-_18007077 | 0.63 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr14_+_11290828 | 0.61 |
ENSDART00000184078
|
rlim
|
ring finger protein, LIM domain interacting |
| chr5_+_62356304 | 0.61 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr23_+_10805188 | 0.61 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr14_-_3944017 | 0.61 |
ENSDART00000055817
|
si:ch73-49o8.1
|
si:ch73-49o8.1 |
| chr11_+_5565082 | 0.61 |
ENSDART00000112590
ENSDART00000183207 |
si:ch73-40i7.5
|
si:ch73-40i7.5 |
| chr1_+_54194801 | 0.60 |
ENSDART00000186802
|
CABZ01067150.1
|
|
| chr16_+_23495600 | 0.60 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr6_-_41021571 | 0.59 |
ENSDART00000103776
|
hdhd3
|
haloacid dehalogenase-like hydrolase domain containing 3 |
| chr6_-_6295513 | 0.59 |
ENSDART00000092172
|
mtif2
|
mitochondrial translational initiation factor 2 |
| chr5_+_1109098 | 0.58 |
ENSDART00000166268
|
LO017790.1
|
|
| chr6_-_31323984 | 0.58 |
ENSDART00000183660
ENSDART00000184146 ENSDART00000180156 |
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr19_+_4968947 | 0.58 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr20_+_32552912 | 0.57 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr11_-_24532988 | 0.56 |
ENSDART00000067078
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr7_+_22399669 | 0.56 |
ENSDART00000144375
|
fgf11a
|
fibroblast growth factor 11a |
| chr5_+_57210237 | 0.56 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr16_+_26735894 | 0.55 |
ENSDART00000191605
ENSDART00000185175 |
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr19_+_19989380 | 0.55 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr13_+_33688474 | 0.54 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr1_-_29761086 | 0.53 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr5_-_30620625 | 0.52 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr23_+_43870886 | 0.51 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr2_+_31806602 | 0.51 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr12_+_26877381 | 0.51 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr5_+_50953240 | 0.51 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr22_+_1786230 | 0.51 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr22_+_35131890 | 0.51 |
ENSDART00000003303
ENSDART00000130581 |
rnf13
|
ring finger protein 13 |
| chr13_+_35690023 | 0.51 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr22_+_11972786 | 0.51 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr16_-_26435431 | 0.49 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr6_-_16393491 | 0.49 |
ENSDART00000179312
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr2_-_26720854 | 0.49 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr17_-_6536305 | 0.48 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr20_+_14789148 | 0.48 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr15_-_15968883 | 0.47 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr1_-_28950366 | 0.47 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr5_+_71924175 | 0.46 |
ENSDART00000115182
ENSDART00000170215 |
nup214
|
nucleoporin 214 |
| chr6_-_55399214 | 0.46 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr16_+_27383717 | 0.46 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr10_+_43406796 | 0.46 |
ENSDART00000184337
ENSDART00000183034 ENSDART00000180623 ENSDART00000132486 |
rasa1b
|
RAS p21 protein activator (GTPase activating protein) 1b |
| chr9_-_23761087 | 0.45 |
ENSDART00000090486
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr7_-_48396193 | 0.45 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr12_+_29173523 | 0.45 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr20_+_41756996 | 0.44 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr22_-_9896180 | 0.43 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr25_-_15268443 | 0.42 |
ENSDART00000151827
|
ccdc73
|
coiled-coil domain containing 73 |
| chr7_+_56889879 | 0.42 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr17_+_21546993 | 0.42 |
ENSDART00000182387
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr24_-_11057305 | 0.42 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr9_-_12886108 | 0.42 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr15_-_6946286 | 0.42 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr9_+_25840720 | 0.40 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr25_+_34247353 | 0.40 |
ENSDART00000148914
|
bnip2
|
BCL2 interacting protein 2 |
| chr20_-_25644131 | 0.40 |
ENSDART00000138997
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr11_+_44503774 | 0.39 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr8_-_14067517 | 0.39 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr4_-_16706776 | 0.39 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr3_-_53092509 | 0.39 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr21_-_2565825 | 0.39 |
ENSDART00000169633
|
col4a3bpb
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein b |
| chr23_+_32029304 | 0.39 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr9_-_5318873 | 0.38 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr16_+_32082547 | 0.38 |
ENSDART00000190122
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr20_+_50052116 | 0.37 |
ENSDART00000047212
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr4_-_13156971 | 0.37 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr5_+_27421025 | 0.37 |
ENSDART00000098590
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr1_-_23157583 | 0.37 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr2_+_25378457 | 0.37 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr1_-_23268569 | 0.37 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr7_-_32875552 | 0.36 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr10_-_11203164 | 0.36 |
ENSDART00000040308
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr2_+_34767171 | 0.36 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr13_+_11828516 | 0.35 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr17_+_6535694 | 0.35 |
ENSDART00000191681
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr19_-_10286228 | 0.35 |
ENSDART00000027316
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr1_-_45614318 | 0.35 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_26906656 | 0.34 |
ENSDART00000156150
|
clec16a
|
C-type lectin domain containing 16A |
| chr22_-_8006342 | 0.34 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr5_+_1055042 | 0.33 |
ENSDART00000046781
|
rnf185
|
ring finger protein 185 |
| chr5_-_6745442 | 0.33 |
ENSDART00000157402
ENSDART00000128684 ENSDART00000168698 |
ostf1
|
osteoclast stimulating factor 1 |
| chr21_-_2958422 | 0.33 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr20_-_26841544 | 0.32 |
ENSDART00000088875
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr22_+_2100774 | 0.32 |
ENSDART00000106436
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr16_+_42471455 | 0.32 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr8_+_47099033 | 0.32 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr3_-_15886658 | 0.32 |
ENSDART00000113630
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr3_-_33997763 | 0.32 |
ENSDART00000151461
ENSDART00000151307 |
ighz
|
immunoglobulin heavy constant zeta |
| chr12_-_26620728 | 0.31 |
ENSDART00000087067
|
hexdc
|
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr2_-_47923803 | 0.31 |
ENSDART00000056306
|
ftr24
|
finTRIM family, member 24 |
| chr21_-_27272657 | 0.31 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr4_-_16706941 | 0.31 |
ENSDART00000033053
ENSDART00000140510 ENSDART00000141119 |
dennd5b
|
DENN/MADD domain containing 5B |
| chr5_-_33825465 | 0.30 |
ENSDART00000110645
|
dab2ipb
|
DAB2 interacting protein b |
| chr19_-_35155722 | 0.30 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr16_+_32729223 | 0.30 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr18_+_9810642 | 0.29 |
ENSDART00000143965
|
CACNA2D1
|
si:dkey-266j7.2 |
| chr21_-_28439596 | 0.29 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr18_-_25276932 | 0.29 |
ENSDART00000076183
|
plin1
|
perilipin 1 |
| chr3_-_11373435 | 0.29 |
ENSDART00000192654
|
AL935044.1
|
|
| chr25_-_4313699 | 0.28 |
ENSDART00000154038
|
syt7a
|
synaptotagmin VIIa |
| chr15_-_8177919 | 0.28 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr10_-_8197049 | 0.28 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr12_+_23912074 | 0.27 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr8_+_53120278 | 0.25 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr20_+_36682051 | 0.24 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr20_+_19562383 | 0.24 |
ENSDART00000103798
|
nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr3_+_35298078 | 0.24 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr11_+_42600731 | 0.23 |
ENSDART00000182753
ENSDART00000192028 ENSDART00000085868 |
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr4_-_25515154 | 0.23 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr6_+_8652310 | 0.23 |
ENSDART00000105098
|
usp40
|
ubiquitin specific peptidase 40 |
| chr11_-_18557277 | 0.22 |
ENSDART00000103950
|
dido1
|
death inducer-obliterator 1 |
| chr3_-_26806032 | 0.22 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_-_8040436 | 0.22 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr19_+_26424734 | 0.22 |
ENSDART00000187402
|
npr1a
|
natriuretic peptide receptor 1a |
| chr5_+_72194444 | 0.22 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr3_-_25492361 | 0.21 |
ENSDART00000147322
ENSDART00000055473 |
grb2b
|
growth factor receptor-bound protein 2b |
| chr7_+_20031202 | 0.20 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr11_+_8152872 | 0.20 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr25_+_388258 | 0.20 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr25_-_18778356 | 0.20 |
ENSDART00000111425
|
fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr21_-_22715297 | 0.19 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr1_-_59313465 | 0.19 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr5_+_25680845 | 0.18 |
ENSDART00000139701
ENSDART00000009952 |
zfand5a
|
zinc finger, AN1-type domain 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.3 | 1.0 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 2.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 1.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.5 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 1.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.0 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 1.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:1902592 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.9 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.5 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.3 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.4 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.7 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.1 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.3 | 1.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 1.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 2.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 1.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 1.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.0 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 0.5 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 0.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.6 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 1.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |