Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
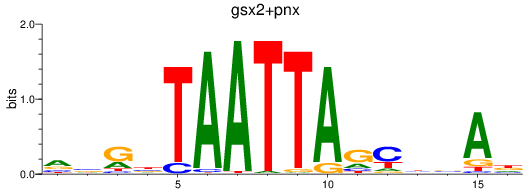
Results for gsx2+pnx
Z-value: 1.92
Transcription factors associated with gsx2+pnx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pnx
|
ENSDARG00000025899 | posterior neuron-specific homeobox |
|
gsx2
|
ENSDARG00000043322 | GS homeobox 2 |
|
gsx2
|
ENSDARG00000116417 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx2 | dr11_v1_chr20_-_22484621_22484621 | 0.85 | 3.9e-03 | Click! |
| pnx | dr11_v1_chr10_+_2799285_2799285 | 0.05 | 8.9e-01 | Click! |
Activity profile of gsx2+pnx motif
Sorted Z-values of gsx2+pnx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22310919 | 7.17 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr25_+_31277415 | 5.41 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr21_+_26390549 | 4.22 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr20_+_26966725 | 3.77 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr25_+_31276842 | 3.40 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr14_-_4145594 | 3.16 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr25_+_29160102 | 2.99 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr15_-_12011390 | 2.87 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr16_+_31804590 | 2.72 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr11_+_38280454 | 2.69 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr23_+_20110086 | 2.58 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr15_+_23799461 | 2.57 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr2_+_55982300 | 2.55 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr12_+_20352400 | 2.51 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr15_+_19646902 | 2.50 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr7_-_48667056 | 2.49 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr21_-_5205617 | 2.22 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr3_-_61217753 | 2.20 |
ENSDART00000182630
|
pvalb5
|
parvalbumin 5 |
| chr17_+_23298928 | 2.20 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr21_+_6751405 | 2.19 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr1_-_59287410 | 2.19 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr21_+_6751760 | 2.15 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr5_+_51597677 | 2.06 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr7_-_24046999 | 2.05 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr25_+_31868268 | 2.04 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr6_-_7720332 | 2.00 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr8_+_29986265 | 1.94 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr15_-_21877726 | 1.93 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr12_+_45677293 | 1.93 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr3_-_46817499 | 1.90 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr13_+_35339182 | 1.90 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr3_+_59784632 | 1.90 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr14_+_11458044 | 1.87 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr1_+_127250 | 1.75 |
ENSDART00000003463
|
f7i
|
coagulation factor VIIi |
| chr11_-_472547 | 1.71 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr25_+_5035343 | 1.71 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr20_+_535458 | 1.70 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr20_+_35382482 | 1.67 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr24_+_3307857 | 1.65 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr13_+_255067 | 1.65 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr11_+_44135351 | 1.65 |
ENSDART00000182914
|
FO704721.1
|
|
| chr8_-_15129573 | 1.64 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr14_-_40390757 | 1.64 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr10_-_26744131 | 1.61 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr4_+_76575585 | 1.61 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr19_-_2861444 | 1.60 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr15_+_33865312 | 1.60 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr5_-_19394440 | 1.60 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr18_-_46256560 | 1.60 |
ENSDART00000171375
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr3_-_23643751 | 1.57 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr1_-_50859053 | 1.56 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr15_-_22074315 | 1.53 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr3_-_18744511 | 1.52 |
ENSDART00000145539
|
zgc:113333
|
zgc:113333 |
| chr3_-_50443607 | 1.52 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr18_+_48423973 | 1.52 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_+_30903816 | 1.51 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr1_-_58036509 | 1.50 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr1_-_58963395 | 1.50 |
ENSDART00000130415
|
CABZ01115881.1
|
|
| chr1_+_50921266 | 1.49 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr20_+_26880668 | 1.49 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr9_-_22834860 | 1.47 |
ENSDART00000146486
|
neb
|
nebulin |
| chr21_-_22827548 | 1.47 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr15_+_1796313 | 1.46 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr6_-_54815886 | 1.46 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr18_-_46258612 | 1.41 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr16_+_11834516 | 1.41 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr3_-_61218272 | 1.41 |
ENSDART00000133091
ENSDART00000055068 |
pvalb5
|
parvalbumin 5 |
| chr19_+_1831911 | 1.41 |
ENSDART00000166653
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr9_-_6372535 | 1.39 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr16_-_42933332 | 1.39 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr7_-_27685365 | 1.38 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr11_+_26476153 | 1.37 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr21_-_37790727 | 1.35 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr16_-_31754102 | 1.35 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_29445473 | 1.35 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr23_+_26979174 | 1.33 |
ENSDART00000018654
|
rnd1b
|
Rho family GTPase 1b |
| chr8_+_52619365 | 1.32 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr10_+_32104305 | 1.31 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr16_+_41826584 | 1.29 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr7_-_30174882 | 1.28 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr13_+_22479988 | 1.26 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr23_+_19813677 | 1.25 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr21_+_18997511 | 1.24 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr10_+_37137464 | 1.22 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr15_-_36533322 | 1.22 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr24_+_22485710 | 1.20 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr2_+_6963296 | 1.19 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr12_+_30265802 | 1.18 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr20_-_9462433 | 1.18 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr19_-_31402429 | 1.18 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr3_+_18398876 | 1.17 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr15_-_4528326 | 1.16 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr11_-_42554290 | 1.15 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr4_+_21129752 | 1.14 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr4_+_22480169 | 1.14 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr21_+_45841731 | 1.13 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr22_+_17205608 | 1.12 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr11_-_30158191 | 1.11 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr1_+_52392511 | 1.11 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr11_-_40728380 | 1.10 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr4_-_73756673 | 1.09 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr20_-_14925281 | 1.09 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr3_-_60827402 | 1.08 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr15_-_35252522 | 1.07 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr3_-_38783951 | 1.06 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr3_+_30500968 | 1.05 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr24_-_38657683 | 1.05 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr5_+_40485503 | 1.05 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr20_-_46362606 | 1.05 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr8_+_1766206 | 1.04 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr21_-_26490186 | 1.04 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr9_+_38372216 | 1.03 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr7_+_38936132 | 1.03 |
ENSDART00000173945
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr25_-_31953516 | 1.02 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr18_-_48547564 | 1.02 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr7_+_8456999 | 1.01 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr14_-_14643190 | 1.01 |
ENSDART00000167119
|
znf185
|
zinc finger protein 185 with LIM domain |
| chr8_-_31107537 | 1.00 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr17_+_6452511 | 1.00 |
ENSDART00000064694
|
TATDN3
|
Danio rerio TatD DNase domain containing 3-like (LOC571912), mRNA. |
| chr15_-_12011202 | 0.98 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr14_-_33824329 | 0.98 |
ENSDART00000189271
ENSDART00000180458 |
vimr2
|
vimentin-related 2 |
| chr20_-_13774826 | 0.97 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr15_-_2632891 | 0.97 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr11_+_7264457 | 0.96 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr8_-_23776399 | 0.96 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr23_-_31512496 | 0.96 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr11_-_41853874 | 0.95 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr15_+_5360407 | 0.95 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr23_+_28582865 | 0.93 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr19_+_43780970 | 0.93 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr21_+_1381276 | 0.92 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr14_+_5385855 | 0.92 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr7_+_69019851 | 0.91 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr19_-_38611814 | 0.91 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr4_+_58576146 | 0.90 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr24_+_6107901 | 0.90 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr1_+_7517454 | 0.88 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr11_-_2131280 | 0.87 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr4_+_47636303 | 0.87 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr1_-_14332283 | 0.87 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr4_+_72723304 | 0.86 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr16_-_25568512 | 0.85 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr11_-_15296805 | 0.85 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr24_-_36680261 | 0.85 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr23_-_969844 | 0.84 |
ENSDART00000127037
|
cdh26.2
|
cadherin 26, tandem duplicate 2 |
| chr13_-_40726865 | 0.83 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr14_-_7137808 | 0.83 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr18_+_5137241 | 0.82 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr12_-_28363111 | 0.80 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr21_+_45757317 | 0.79 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr4_-_45100253 | 0.79 |
ENSDART00000163870
|
si:dkey-51d8.3
|
si:dkey-51d8.3 |
| chr24_-_6078222 | 0.79 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr4_-_77506362 | 0.79 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr23_-_44219902 | 0.79 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr7_+_30787903 | 0.78 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr14_+_30795559 | 0.77 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr24_-_37640705 | 0.77 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr3_-_61099004 | 0.77 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr22_-_12746539 | 0.76 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr12_+_22580579 | 0.76 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_-_5669122 | 0.75 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr9_-_5263947 | 0.74 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr3_-_30625219 | 0.74 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr6_+_6924637 | 0.74 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr7_-_71758307 | 0.74 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr14_+_924876 | 0.73 |
ENSDART00000183908
|
myoz3a
|
myozenin 3a |
| chr22_-_8388678 | 0.73 |
ENSDART00000184389
|
CABZ01061498.1
|
|
| chr20_-_14924858 | 0.72 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr24_-_31425799 | 0.72 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr21_-_42097736 | 0.71 |
ENSDART00000100000
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr18_+_1154189 | 0.71 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr3_-_34084387 | 0.71 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr8_-_24803111 | 0.71 |
ENSDART00000186281
|
BX324132.3
|
|
| chr20_-_35578435 | 0.70 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr3_-_23407720 | 0.70 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr20_+_25225112 | 0.70 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr18_+_23408073 | 0.68 |
ENSDART00000136489
|
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr14_-_36320506 | 0.67 |
ENSDART00000074639
|
egf
|
epidermal growth factor |
| chr25_+_469855 | 0.67 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr3_-_58798377 | 0.66 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr2_+_33382648 | 0.65 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_-_51288178 | 0.65 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr18_-_2433011 | 0.64 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr19_-_7495006 | 0.63 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr6_-_30954268 | 0.62 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr15_-_2493771 | 0.62 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr25_-_13839743 | 0.62 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr5_+_1079423 | 0.61 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr23_+_40460333 | 0.60 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr14_+_22591624 | 0.59 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr19_-_657439 | 0.58 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr5_-_69649044 | 0.57 |
ENSDART00000011192
ENSDART00000170885 |
eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha |
| chr7_+_54642005 | 0.56 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr18_+_11506561 | 0.56 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr11_-_21528056 | 0.55 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr24_+_11908833 | 0.54 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr5_-_61624693 | 0.54 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr3_+_34919810 | 0.53 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr21_-_45076778 | 0.53 |
ENSDART00000181525
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr23_-_1658980 | 0.52 |
ENSDART00000186227
|
CU693481.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx2+pnx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.5 | 1.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 1.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.4 | 1.9 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.4 | 2.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 3.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.3 | 1.7 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 0.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.3 | 1.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 1.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 13.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 0.8 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.2 | 1.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 1.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.7 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.7 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.0 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.2 | 0.9 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 3.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 0.6 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 2.1 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.3 | GO:0046635 | positive regulation of alpha-beta T cell activation(GO:0046635) |
| 0.1 | 1.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.5 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 1.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 2.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 1.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 1.0 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 1.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.5 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 3.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 2.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.3 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 7.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.5 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.9 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 4.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.2 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 3.0 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.3 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 1.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.6 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 2.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.9 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.7 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 3.0 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 4.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 18.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.4 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.5 | 1.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.4 | 1.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.4 | 2.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 1.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 2.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 0.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 3.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 2.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 3.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.4 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 9.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.7 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 2.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.1 | 2.0 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.3 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 2.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 4.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 2.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 13.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.0 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 7.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 3.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME TRANSLATION | Genes involved in Translation |