Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
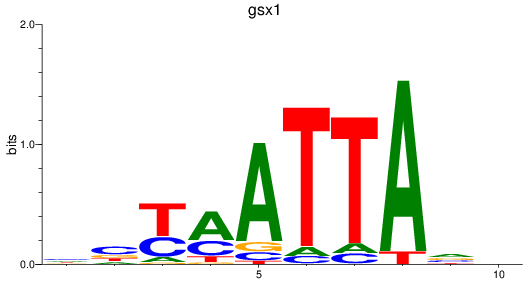
Results for gsx1
Z-value: 1.24
Transcription factors associated with gsx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsx1
|
ENSDARG00000035735 | GS homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx1 | dr11_v1_chr5_-_67911111_67911111 | 0.87 | 2.6e-03 | Click! |
Activity profile of gsx1 motif
Sorted Z-values of gsx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 4.30 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr7_+_20503344 | 3.03 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr15_+_23799461 | 3.01 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr21_+_28445052 | 2.71 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr3_+_46635527 | 2.40 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr8_+_21353878 | 2.21 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr25_+_14507567 | 2.17 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr19_-_31402429 | 1.98 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr9_-_21918963 | 1.97 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr14_+_45406299 | 1.73 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr15_-_21877726 | 1.64 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr16_+_5774977 | 1.63 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr11_-_45138857 | 1.59 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_+_46356879 | 1.59 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr22_-_10121880 | 1.58 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr15_-_23376541 | 1.54 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr24_-_6078222 | 1.53 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr9_-_32753535 | 1.44 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr19_+_43297546 | 1.42 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr12_+_20352400 | 1.41 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr3_-_25814097 | 1.41 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr20_+_19512727 | 1.35 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr1_+_34696503 | 1.33 |
ENSDART00000186106
|
CR339054.2
|
|
| chr14_-_17072736 | 1.31 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr11_+_10909183 | 1.31 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr8_+_23738122 | 1.30 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr21_-_23110841 | 1.26 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr16_-_43344859 | 1.26 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr25_-_5119162 | 1.24 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr20_+_52554352 | 1.23 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr18_+_24921587 | 1.21 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr24_+_9412450 | 1.21 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr15_-_1822548 | 1.19 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr13_+_27232848 | 1.18 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr21_-_37973819 | 1.17 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr15_-_22074315 | 1.16 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr25_+_5068442 | 1.15 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr12_+_3078221 | 1.15 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr9_+_25776971 | 1.14 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr9_-_6372535 | 1.13 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr14_-_4273396 | 1.12 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr19_-_42571829 | 1.11 |
ENSDART00000102606
|
zgc:103438
|
zgc:103438 |
| chr16_+_31804590 | 1.08 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr15_+_1796313 | 1.08 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr2_+_16160906 | 1.07 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr10_+_16584382 | 1.06 |
ENSDART00000112039
|
CR790388.1
|
|
| chr5_+_27897504 | 1.05 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr16_-_28878080 | 1.02 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr8_-_25336589 | 1.01 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr21_-_22827548 | 0.99 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr1_+_55293424 | 0.98 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr19_+_14921000 | 0.97 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr10_+_31646020 | 0.97 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr4_-_9891874 | 0.95 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr19_-_42556086 | 0.93 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr3_+_46459540 | 0.89 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr9_+_21146862 | 0.88 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr5_-_33287691 | 0.87 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr8_-_18537866 | 0.86 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr2_-_11512819 | 0.83 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr17_+_53425037 | 0.82 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr24_-_40744672 | 0.82 |
ENSDART00000160672
|
CU633479.1
|
|
| chr6_-_46941245 | 0.80 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr19_+_10339538 | 0.80 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr20_+_10723292 | 0.79 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr15_+_36309070 | 0.79 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr23_+_40460333 | 0.78 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr13_+_29238850 | 0.74 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr25_+_32755485 | 0.73 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr8_+_24747865 | 0.73 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr13_+_28903959 | 0.73 |
ENSDART00000166435
|
si:ch73-28h20.1
|
si:ch73-28h20.1 |
| chr23_+_11285662 | 0.72 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr6_+_39232245 | 0.72 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr1_-_50859053 | 0.71 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr8_+_30452945 | 0.71 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr24_-_37640705 | 0.70 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr25_-_6011034 | 0.70 |
ENSDART00000075197
ENSDART00000136054 |
snx22
|
sorting nexin 22 |
| chr9_-_20372977 | 0.70 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr8_-_17167819 | 0.69 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr7_+_69653981 | 0.68 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr6_+_53349966 | 0.68 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr6_-_32999646 | 0.67 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr24_+_6107901 | 0.66 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr4_+_5798223 | 0.65 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr4_+_11479705 | 0.65 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr10_+_21867307 | 0.64 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr17_-_23416897 | 0.63 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr7_+_756942 | 0.62 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr9_-_31278048 | 0.62 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr21_-_17296789 | 0.62 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr9_-_52962521 | 0.61 |
ENSDART00000170419
|
CU855885.1
|
|
| chr8_+_46641314 | 0.60 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr8_+_23521974 | 0.57 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr11_-_29737088 | 0.57 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr13_-_20381485 | 0.57 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr8_-_25120231 | 0.57 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr3_+_32553714 | 0.56 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr18_+_14564085 | 0.55 |
ENSDART00000009363
ENSDART00000141813 ENSDART00000136120 |
si:dkey-246g23.4
|
si:dkey-246g23.4 |
| chr5_+_37840914 | 0.55 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr2_-_23768818 | 0.54 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr5_-_21970881 | 0.54 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr4_+_52356485 | 0.53 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr1_-_10647307 | 0.53 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr14_+_30795559 | 0.52 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr20_-_46362606 | 0.52 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr24_+_17007407 | 0.52 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr6_-_46861676 | 0.52 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr22_-_11137268 | 0.52 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr21_-_40174647 | 0.52 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr8_-_23612462 | 0.50 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr22_+_20427170 | 0.50 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr14_-_21959712 | 0.50 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr2_-_10600950 | 0.49 |
ENSDART00000160216
|
BX323564.1
|
|
| chr11_+_30057762 | 0.49 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr11_+_36665359 | 0.48 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr12_+_10631266 | 0.48 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr1_-_10647484 | 0.48 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr2_-_59285085 | 0.48 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr18_-_6982499 | 0.47 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr1_+_33668236 | 0.46 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr7_+_2236317 | 0.46 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr3_-_29941357 | 0.45 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr3_-_32169754 | 0.45 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr1_+_55239160 | 0.45 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr13_+_35637048 | 0.45 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr19_-_30524952 | 0.45 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr22_-_12746539 | 0.44 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr15_+_9053059 | 0.44 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr17_+_46818521 | 0.42 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr1_-_9123465 | 0.40 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr15_-_19705707 | 0.39 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
| chr25_+_20715950 | 0.39 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr5_-_44286987 | 0.38 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr25_-_26758253 | 0.38 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr8_+_8936912 | 0.37 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr11_-_42980535 | 0.37 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr4_-_51460013 | 0.36 |
ENSDART00000193382
|
CR628395.1
|
|
| chr19_+_2279051 | 0.35 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr23_+_31913292 | 0.34 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr20_-_52902693 | 0.34 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr14_+_7377552 | 0.33 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr15_-_25518084 | 0.33 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr8_-_50888806 | 0.33 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr16_-_24605969 | 0.33 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr19_-_12315693 | 0.33 |
ENSDART00000151158
|
ncaldb
|
neurocalcin delta b |
| chr22_-_8725768 | 0.32 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr4_-_71210030 | 0.32 |
ENSDART00000186473
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr2_+_3986083 | 0.31 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr14_+_33329761 | 0.31 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr14_+_22022441 | 0.31 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr4_-_42408339 | 0.30 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr19_+_46237665 | 0.29 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr1_+_31110817 | 0.29 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr15_+_34988148 | 0.29 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr2_+_33326522 | 0.28 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr25_+_16689633 | 0.28 |
ENSDART00000073416
|
ada2a
|
adenosine deaminase 2a |
| chr8_+_48484455 | 0.28 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr15_-_31323932 | 0.27 |
ENSDART00000132455
|
or118-2
|
odorant receptor, family F, subfamily 118, member 2 |
| chr17_-_45386546 | 0.27 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr21_-_45077429 | 0.26 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr2_+_19578446 | 0.26 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr20_-_47188966 | 0.26 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr4_+_9400012 | 0.26 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr10_+_2799285 | 0.25 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr11_+_14284866 | 0.25 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr20_+_42978499 | 0.24 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr13_+_15190677 | 0.23 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr20_-_35040041 | 0.22 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr2_-_51511494 | 0.22 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr9_+_23224761 | 0.22 |
ENSDART00000142008
|
map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr22_-_36530902 | 0.20 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_+_29462249 | 0.20 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr9_+_41156818 | 0.20 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr3_+_1637358 | 0.19 |
ENSDART00000180266
|
CR394546.5
|
|
| chr18_+_7073130 | 0.19 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr22_+_10660140 | 0.19 |
ENSDART00000105835
ENSDART00000038511 |
tusc2b
|
tumor suppressor candidate 2b |
| chr11_+_24703108 | 0.19 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr4_+_9177997 | 0.18 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr25_-_27722614 | 0.18 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr8_-_13184989 | 0.17 |
ENSDART00000135738
|
zgc:194990
|
zgc:194990 |
| chr20_+_39457598 | 0.17 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr10_+_40633990 | 0.16 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr8_+_23355484 | 0.16 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr21_-_7035599 | 0.16 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr22_-_10586191 | 0.16 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr23_+_404975 | 0.15 |
ENSDART00000181336
|
CU929146.1
|
|
| chr10_+_40756352 | 0.15 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr25_+_28825657 | 0.14 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr6_+_41191482 | 0.13 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chrM_+_9735 | 0.13 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr11_+_36231248 | 0.12 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr7_+_38509333 | 0.12 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr8_+_25034544 | 0.12 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr12_+_16168342 | 0.12 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr6_-_40581376 | 0.11 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr15_-_41457898 | 0.11 |
ENSDART00000188847
|
nlrc9
|
NLR family CARD domain containing 9 |
| chr21_+_43404945 | 0.10 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr3_-_17716322 | 0.09 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr4_+_43246676 | 0.09 |
ENSDART00000187588
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr21_+_35041399 | 0.09 |
ENSDART00000188183
ENSDART00000021645 |
kdelc2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.5 | 1.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.4 | 1.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 1.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.3 | 1.6 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.3 | 1.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 1.6 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 0.8 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 3.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.4 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.5 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 4.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 1.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 2.2 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.3 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.7 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 1.2 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.7 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 4.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.2 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 2.7 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.0 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 1.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.0 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |