Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
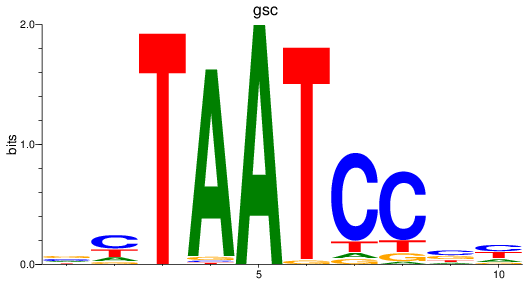
Results for gsc
Z-value: 1.11
Transcription factors associated with gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsc
|
ENSDARG00000059073 | goosecoid |
|
gsc
|
ENSDARG00000111184 | goosecoid |
|
gsc
|
ENSDARG00000115937 | goosecoid |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsc | dr11_v1_chr17_-_19345521_19345521 | 0.43 | 2.5e-01 | Click! |
Activity profile of gsc motif
Sorted Z-values of gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_54077740 | 2.45 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr11_-_15090118 | 1.36 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr11_-_15090564 | 1.24 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr7_-_13882988 | 1.13 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr14_-_36378494 | 1.13 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr14_+_46313396 | 1.11 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_-_345503 | 1.03 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr14_+_46313135 | 1.02 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_+_41186320 | 1.01 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr17_-_51202339 | 0.99 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr14_+_32838110 | 0.88 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr22_+_34701848 | 0.88 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr5_+_28973264 | 0.83 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr5_+_28972935 | 0.81 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr3_-_28828242 | 0.74 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr3_+_26144765 | 0.71 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr19_+_40856534 | 0.70 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_+_32837914 | 0.70 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr10_-_20453995 | 0.67 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr14_+_32839535 | 0.66 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr15_+_45640906 | 0.62 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr16_+_32059785 | 0.59 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr22_-_910926 | 0.58 |
ENSDART00000180075
|
FP016205.1
|
|
| chr8_+_23142946 | 0.56 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr5_-_28625515 | 0.56 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr22_+_5103349 | 0.56 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr17_-_16965809 | 0.55 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr19_+_40856807 | 0.54 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr9_+_32978302 | 0.52 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr6_+_48618512 | 0.51 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr10_-_22845485 | 0.51 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr11_-_44873780 | 0.51 |
ENSDART00000160465
|
opn6a
|
opsin 6, group member a |
| chr4_-_7212875 | 0.48 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr5_+_1278092 | 0.47 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr15_+_9053059 | 0.46 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr19_+_24882845 | 0.45 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr13_-_31622195 | 0.45 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr7_-_57933736 | 0.44 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr25_+_22274642 | 0.43 |
ENSDART00000127099
|
nr2e3
|
nuclear receptor subfamily 2, group E, member 3 |
| chr7_-_35516251 | 0.43 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr11_+_36180349 | 0.43 |
ENSDART00000012940
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr1_-_7894255 | 0.40 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr13_+_3954715 | 0.40 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr6_-_35401282 | 0.39 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr20_+_37295006 | 0.39 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr6_+_9867426 | 0.39 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr17_-_32863250 | 0.39 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr2_-_5356686 | 0.39 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr18_+_49969568 | 0.38 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr3_+_34919810 | 0.38 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr17_+_36627099 | 0.38 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr13_+_3954540 | 0.37 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr25_+_192116 | 0.37 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr7_-_49594995 | 0.37 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr1_+_47585700 | 0.37 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr9_-_29544720 | 0.36 |
ENSDART00000130317
|
arhgap20
|
Rho GTPase activating protein 20 |
| chr20_-_18736281 | 0.36 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr25_-_32751982 | 0.36 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr18_-_1228688 | 0.35 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr2_-_52365251 | 0.35 |
ENSDART00000097716
|
tle2c
|
transducin like enhancer of split 2c |
| chr24_-_6501211 | 0.35 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr2_-_4797512 | 0.34 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr3_-_30941362 | 0.34 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr20_-_25436389 | 0.33 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr23_+_6232895 | 0.32 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr5_-_38161033 | 0.32 |
ENSDART00000145907
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr24_-_24060632 | 0.32 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr13_+_29462249 | 0.32 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr2_-_1569250 | 0.31 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr2_+_30721070 | 0.31 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr19_-_41069573 | 0.30 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr1_-_7893808 | 0.30 |
ENSDART00000110154
|
radil
|
Ras association and DIL domains |
| chr18_-_36135799 | 0.30 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr2_-_30784198 | 0.30 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr7_-_18656069 | 0.29 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr22_+_661505 | 0.29 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr24_-_7777389 | 0.29 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr25_+_19041329 | 0.29 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr19_-_10373630 | 0.28 |
ENSDART00000131517
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr7_+_30970045 | 0.27 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr20_-_9963713 | 0.26 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr9_+_13685921 | 0.26 |
ENSDART00000145775
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr21_-_28235361 | 0.26 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr20_-_14680897 | 0.26 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr22_+_38581012 | 0.25 |
ENSDART00000169239
|
BX088728.1
|
|
| chr10_-_36738619 | 0.24 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr12_-_10705916 | 0.24 |
ENSDART00000164038
|
FO704786.1
|
|
| chr17_-_20979077 | 0.23 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr19_+_2275019 | 0.23 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr23_-_21797517 | 0.23 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr7_-_35515931 | 0.22 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr2_-_31664615 | 0.22 |
ENSDART00000190546
ENSDART00000111810 |
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr1_+_47091468 | 0.22 |
ENSDART00000036783
|
cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr20_+_53441935 | 0.22 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr6_-_19270484 | 0.21 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr2_+_30878864 | 0.21 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr18_+_19975787 | 0.21 |
ENSDART00000138103
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr1_-_45920632 | 0.20 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr18_-_21640389 | 0.20 |
ENSDART00000100857
|
slc38a8a
|
solute carrier family 38, member 8a |
| chr3_+_52475058 | 0.20 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr22_+_661711 | 0.20 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr5_-_14390445 | 0.20 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr2_+_28995776 | 0.20 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr10_-_22249444 | 0.20 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr17_-_15657029 | 0.20 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr9_+_33357011 | 0.20 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr7_-_50517023 | 0.20 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr2_+_24199073 | 0.19 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr17_+_15845765 | 0.19 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr5_+_43072951 | 0.19 |
ENSDART00000193905
ENSDART00000026020 |
wdr54
|
WD repeat domain 54 |
| chr22_+_38164486 | 0.19 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr13_+_12528043 | 0.18 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr15_+_15375607 | 0.18 |
ENSDART00000129751
|
si:ch211-105f12.2
|
si:ch211-105f12.2 |
| chr1_+_16573982 | 0.17 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr16_-_17188294 | 0.17 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr3_+_23703704 | 0.16 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr21_-_39327223 | 0.16 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr12_+_41991635 | 0.16 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr23_-_5759242 | 0.16 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr2_-_30055432 | 0.14 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr16_+_25095483 | 0.14 |
ENSDART00000155032
|
si:ch211-261d7.3
|
si:ch211-261d7.3 |
| chr3_+_34220194 | 0.14 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr14_-_29799993 | 0.14 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr15_-_38154616 | 0.14 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr25_+_27923846 | 0.14 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr5_-_33274645 | 0.13 |
ENSDART00000188584
|
kyat1
|
kynurenine aminotransferase 1 |
| chr2_-_9818640 | 0.13 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr8_+_48848200 | 0.13 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr10_+_32104305 | 0.13 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr19_+_18799319 | 0.13 |
ENSDART00000171843
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_+_33035709 | 0.13 |
ENSDART00000131660
|
angptl2b
|
angiopoietin-like 2b |
| chr3_-_58798815 | 0.12 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr18_-_17531483 | 0.11 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr23_-_17509656 | 0.11 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr10_+_13279079 | 0.11 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr19_+_18797623 | 0.11 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr24_-_24060460 | 0.11 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr3_-_58798377 | 0.10 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr18_-_6943577 | 0.10 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr3_+_49521106 | 0.10 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr23_-_28294763 | 0.09 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr12_+_47044707 | 0.09 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr24_-_38079261 | 0.09 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr5_-_57526807 | 0.09 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr3_+_23742868 | 0.09 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr12_-_10038870 | 0.09 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr10_+_29260096 | 0.08 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr1_-_45177373 | 0.08 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr12_+_5530247 | 0.08 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr17_-_15149192 | 0.08 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr4_-_14926637 | 0.08 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr11_-_29563437 | 0.08 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr22_-_26005894 | 0.08 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr7_+_4384863 | 0.08 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr16_+_42830152 | 0.08 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr19_-_32944050 | 0.08 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr10_-_37075361 | 0.07 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr12_+_7865470 | 0.07 |
ENSDART00000161683
|
BX548028.1
|
|
| chr21_-_36948 | 0.07 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_-_28148033 | 0.07 |
ENSDART00000177182
|
lactbl1b
|
lactamase, beta-like 1b |
| chr22_-_26100282 | 0.07 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr23_+_13814978 | 0.07 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr7_-_66864756 | 0.06 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr3_-_16227683 | 0.06 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr21_-_20733615 | 0.06 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr24_-_1303553 | 0.05 |
ENSDART00000190984
|
nrp1a
|
neuropilin 1a |
| chr25_-_17552299 | 0.05 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr3_+_26322596 | 0.05 |
ENSDART00000172700
|
si:ch211-156b7.5
|
si:ch211-156b7.5 |
| chr9_+_10770880 | 0.05 |
ENSDART00000003543
ENSDART00000178966 |
gmppaa
|
GDP-mannose pyrophosphorylase Aa |
| chr7_-_12821277 | 0.04 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr23_-_1587955 | 0.04 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr10_-_7988396 | 0.04 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr9_-_48296469 | 0.04 |
ENSDART00000058255
|
bbs5
|
Bardet-Biedl syndrome 5 |
| chr16_+_29555801 | 0.04 |
ENSDART00000169425
|
ensab
|
endosulfine alpha b |
| chr10_-_41400049 | 0.03 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr10_-_22506044 | 0.03 |
ENSDART00000100497
|
zgc:172339
|
zgc:172339 |
| chr3_+_23743139 | 0.03 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr3_+_14157090 | 0.03 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr4_-_76484737 | 0.02 |
ENSDART00000183816
|
ftr51
|
finTRIM family, member 51 |
| chr5_+_67371650 | 0.02 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr25_-_27819838 | 0.02 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr21_-_15046065 | 0.02 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr17_-_33414781 | 0.02 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr19_+_1097393 | 0.02 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr5_+_24287927 | 0.02 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr15_-_14486534 | 0.02 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr16_+_42829735 | 0.02 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr9_+_20519846 | 0.02 |
ENSDART00000109680
ENSDART00000142220 |
vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr19_+_35799384 | 0.01 |
ENSDART00000076023
|
angpt2b
|
angiopoietin 2b |
| chr3_+_46764278 | 0.01 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr11_+_27973055 | 0.01 |
ENSDART00000135135
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr3_-_16227490 | 0.01 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr7_+_41303572 | 0.01 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr6_-_39275793 | 0.00 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr2_+_4146606 | 0.00 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 2.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.2 | 0.6 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.7 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.0 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.5 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0060221 | pineal gland development(GO:0021982) retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 1.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 1.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 1.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 3.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 1.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 0.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |