Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
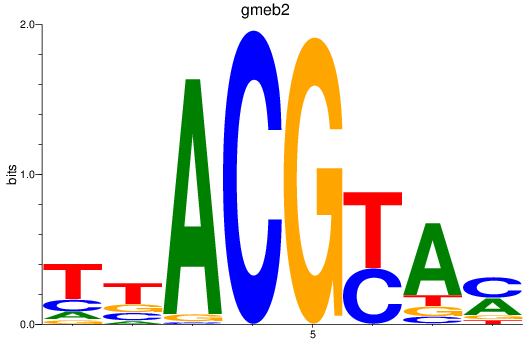
Results for gmeb2
Z-value: 0.72
Transcription factors associated with gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb2
|
ENSDARG00000093240 | si_ch73-302a13.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | dr11_v1_chr23_-_41824460_41824460 | 0.91 | 5.8e-04 | Click! |
Activity profile of gmeb2 motif
Sorted Z-values of gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_29910547 | 1.53 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr17_-_51818659 | 1.22 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr24_+_34069675 | 1.21 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr1_-_49521407 | 1.20 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr15_-_20412286 | 1.13 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr17_-_36860988 | 1.06 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr2_-_17114852 | 1.05 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_17115256 | 1.04 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr13_+_40501455 | 0.99 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr18_-_20674121 | 0.96 |
ENSDART00000005145
|
asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr15_+_5116179 | 0.92 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr13_+_42309688 | 0.91 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr17_-_25831569 | 0.83 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr21_+_18405585 | 0.83 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr8_-_38022298 | 0.79 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr3_-_46410387 | 0.78 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr7_-_37895771 | 0.77 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr19_+_19241372 | 0.76 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr11_+_11120532 | 0.75 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr7_+_13824150 | 0.71 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr20_-_44576949 | 0.68 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr24_+_37484661 | 0.66 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr21_+_1647990 | 0.65 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr3_+_53156813 | 0.65 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr2_-_21170517 | 0.64 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr3_+_32789605 | 0.63 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr10_+_16501699 | 0.61 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr20_-_31743553 | 0.60 |
ENSDART00000087405
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr7_+_13382852 | 0.59 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr1_+_23274710 | 0.59 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr4_+_9011825 | 0.59 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr10_+_6884123 | 0.58 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr21_-_33995710 | 0.56 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr8_+_30112655 | 0.56 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr12_+_17603528 | 0.56 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr17_-_12058171 | 0.55 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr19_+_46259619 | 0.54 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr3_+_22335030 | 0.54 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr5_-_69944084 | 0.53 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr21_+_6397463 | 0.53 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr22_-_817479 | 0.52 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr5_-_13076779 | 0.52 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr14_-_22495604 | 0.52 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr5_-_54395488 | 0.51 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr19_-_432083 | 0.51 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr13_-_36680531 | 0.51 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr3_-_11089062 | 0.50 |
ENSDART00000168938
|
mif4gdb
|
MIF4G domain containing b |
| chr20_+_25486021 | 0.49 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr2_-_54387550 | 0.49 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr6_-_60079551 | 0.48 |
ENSDART00000154753
|
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr18_-_14879135 | 0.48 |
ENSDART00000099701
|
selenoo1
|
selenoprotein O1 |
| chr10_-_16868211 | 0.48 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr23_+_17522867 | 0.48 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr4_+_9011448 | 0.47 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr23_-_35790235 | 0.47 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr20_-_32405440 | 0.46 |
ENSDART00000062978
ENSDART00000153411 |
afg1lb
|
AFG1 like ATPase b |
| chr11_-_18799827 | 0.45 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr23_-_33738570 | 0.45 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr9_-_4598883 | 0.44 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr12_+_11650146 | 0.44 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr7_+_69528850 | 0.44 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr23_-_33558161 | 0.43 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr6_-_24053404 | 0.42 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr8_-_5267442 | 0.42 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr20_-_37933237 | 0.42 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr12_+_28956374 | 0.41 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr10_+_43088270 | 0.41 |
ENSDART00000191221
|
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr5_-_51198430 | 0.41 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr10_+_29137482 | 0.41 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr8_-_32354677 | 0.41 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr10_-_41352502 | 0.41 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr5_+_57210237 | 0.40 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr23_+_2703044 | 0.39 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr7_+_52761841 | 0.39 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr5_+_26138313 | 0.39 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr10_+_29138021 | 0.39 |
ENSDART00000025227
ENSDART00000123033 ENSDART00000034242 |
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr22_-_547748 | 0.38 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr21_-_44772738 | 0.38 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr10_-_21941443 | 0.38 |
ENSDART00000174954
|
FO744833.1
|
|
| chr12_-_9132682 | 0.38 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr8_-_19467011 | 0.37 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr14_+_10625112 | 0.37 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr18_+_5917625 | 0.36 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr23_-_33738945 | 0.36 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr15_-_11683529 | 0.36 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr25_+_21098675 | 0.35 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr13_+_36680564 | 0.35 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr3_-_13146631 | 0.35 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr8_-_1219815 | 0.35 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr20_+_25486206 | 0.35 |
ENSDART00000172076
|
hook1
|
hook microtubule-tethering protein 1 |
| chr14_+_8947282 | 0.35 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr14_-_8890437 | 0.34 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr9_+_25840720 | 0.34 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr25_+_36045072 | 0.34 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr7_+_51795667 | 0.33 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr13_+_30035253 | 0.33 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr7_+_55292959 | 0.33 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr19_-_20163197 | 0.32 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr13_+_2442841 | 0.32 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr17_-_6076266 | 0.32 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr2_+_38055529 | 0.32 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr12_+_33460794 | 0.32 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr1_+_52662203 | 0.31 |
ENSDART00000141530
|
osbp
|
oxysterol binding protein |
| chr20_-_31743817 | 0.31 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr16_+_40131473 | 0.30 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr25_+_21098990 | 0.30 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr6_-_37745508 | 0.30 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr9_+_12948511 | 0.29 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr3_+_46764278 | 0.29 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_38570878 | 0.29 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr17_-_31611692 | 0.29 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr1_+_26105141 | 0.29 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr8_+_47897734 | 0.29 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr3_+_32933663 | 0.29 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr5_-_24245218 | 0.29 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr3_-_13147310 | 0.29 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr8_+_13064750 | 0.28 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr24_+_5208171 | 0.28 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr12_-_27588299 | 0.28 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr25_-_21066136 | 0.27 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr17_-_6076084 | 0.26 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr3_-_30186296 | 0.26 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr7_+_66634167 | 0.25 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr2_+_10821127 | 0.25 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr2_+_10821579 | 0.23 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr1_-_31516091 | 0.23 |
ENSDART00000139828
ENSDART00000146567 |
cenpk
|
centromere protein K |
| chr1_+_49415281 | 0.23 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_-_47427016 | 0.23 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr13_+_21676235 | 0.23 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr17_-_37195163 | 0.23 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr15_+_32798333 | 0.23 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr12_+_20552190 | 0.23 |
ENSDART00000113224
|
gna13a
|
guanine nucleotide binding protein (G protein), alpha 13a |
| chr7_-_39751540 | 0.23 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr18_+_49248389 | 0.22 |
ENSDART00000059285
ENSDART00000142004 ENSDART00000132751 |
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr6_-_39903393 | 0.22 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr23_-_4975452 | 0.22 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr2_-_3403020 | 0.22 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr25_-_17494364 | 0.22 |
ENSDART00000154134
|
lrrc29
|
leucine rich repeat containing 29 |
| chr24_-_7826489 | 0.21 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr8_+_20455134 | 0.21 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr17_-_43399896 | 0.21 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr25_-_2723682 | 0.21 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr20_+_38837238 | 0.20 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr22_-_22301672 | 0.20 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_54387710 | 0.19 |
ENSDART00000127124
|
rab12
|
RAB12, member RAS oncogene family |
| chr3_+_46764022 | 0.19 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr14_-_21932403 | 0.19 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr4_+_23126558 | 0.19 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr2_-_32826108 | 0.18 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr25_-_6389713 | 0.18 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr2_+_52049239 | 0.18 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr6_-_40455109 | 0.18 |
ENSDART00000103878
|
vhl
|
von Hippel-Lindau tumor suppressor |
| chr15_-_23692359 | 0.18 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr22_-_22301929 | 0.18 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr15_-_43284021 | 0.18 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr3_-_8765165 | 0.17 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr5_-_67365006 | 0.17 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr5_-_6567464 | 0.16 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr2_+_32826235 | 0.16 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr2_-_32486080 | 0.16 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr19_-_35155722 | 0.16 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr16_-_33817828 | 0.16 |
ENSDART00000188395
|
rspo1
|
R-spondin 1 |
| chr9_+_19529951 | 0.15 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr13_+_36923052 | 0.15 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr9_-_7655243 | 0.15 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr17_-_49407091 | 0.15 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr2_+_2168547 | 0.14 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr10_+_28428222 | 0.14 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr11_+_29770966 | 0.13 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr5_-_29238889 | 0.13 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr21_-_11856143 | 0.13 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr6_+_9163007 | 0.13 |
ENSDART00000183054
ENSDART00000157552 |
zgc:112023
|
zgc:112023 |
| chr25_-_28668776 | 0.13 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr3_+_46763745 | 0.12 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr9_+_24920677 | 0.12 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr5_-_23596339 | 0.12 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr6_-_51541488 | 0.12 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr22_+_8800432 | 0.11 |
ENSDART00000139940
|
si:dkey-182g1.6
|
si:dkey-182g1.6 |
| chr8_+_23390844 | 0.11 |
ENSDART00000184556
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr2_+_56012016 | 0.10 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr13_-_293250 | 0.10 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr20_-_9199721 | 0.10 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr8_-_4760723 | 0.10 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr13_+_24584401 | 0.10 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr8_+_53311965 | 0.09 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr10_-_15910974 | 0.09 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr21_-_11855828 | 0.09 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr12_+_3571770 | 0.09 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr3_+_59880317 | 0.09 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr6_-_10728057 | 0.09 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr11_+_24298432 | 0.08 |
ENSDART00000138487
|
si:dkey-76p14.2
|
si:dkey-76p14.2 |
| chr11_-_41132296 | 0.08 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr22_+_24214665 | 0.07 |
ENSDART00000163980
ENSDART00000167996 |
glrx2
|
glutaredoxin 2 |
| chr13_+_3938526 | 0.07 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr6_-_17849786 | 0.07 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr18_+_31410652 | 0.07 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr23_+_20640484 | 0.06 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr7_+_41313568 | 0.06 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr12_+_45238292 | 0.06 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr22_-_11493236 | 0.05 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 1.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 0.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 2.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.6 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.3 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.8 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.2 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.6 | GO:0008406 | gonad development(GO:0008406) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.6 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 1.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.8 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 2.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.9 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 0.5 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 1.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.2 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |