Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
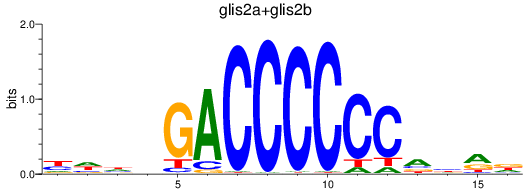
Results for glis2a+glis2b
Z-value: 0.45
Transcription factors associated with glis2a+glis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis2a
|
ENSDARG00000078388 | GLIS family zinc finger 2a |
|
glis2b
|
ENSDARG00000100232 | GLIS family zinc finger 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| glis2b | dr11_v1_chr3_+_12334509_12334509 | 0.68 | 4.5e-02 | Click! |
| glis2a | dr11_v1_chr22_-_26558166_26558166 | 0.01 | 9.8e-01 | Click! |
Activity profile of glis2a+glis2b motif
Sorted Z-values of glis2a+glis2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_31804481 | 1.40 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr20_+_25340814 | 0.88 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr12_-_4070058 | 0.79 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr7_+_18364176 | 0.79 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr23_-_17470146 | 0.68 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr7_+_57725708 | 0.66 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr15_-_97205 | 0.60 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr4_-_17725008 | 0.43 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr25_+_13191391 | 0.41 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_+_21701568 | 0.39 |
ENSDART00000090748
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr19_-_15281996 | 0.37 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr12_-_41619257 | 0.37 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr25_+_13191615 | 0.35 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_-_13116337 | 0.35 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr16_-_16590780 | 0.33 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr5_-_42272517 | 0.32 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_-_45705525 | 0.30 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr24_+_25471196 | 0.28 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr17_-_20717845 | 0.27 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr3_-_61592417 | 0.25 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr16_+_3004422 | 0.24 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr23_-_29004753 | 0.23 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr10_+_33982010 | 0.22 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr23_-_29003864 | 0.22 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr17_-_3291369 | 0.20 |
ENSDART00000181840
|
CABZ01007222.1
|
|
| chr24_-_9960290 | 0.20 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr12_+_2676878 | 0.20 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr13_-_638485 | 0.19 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr5_-_71995108 | 0.18 |
ENSDART00000124587
|
fam78ab
|
family with sequence similarity 78, member Ab |
| chr21_+_25643880 | 0.16 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr2_+_51783120 | 0.16 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr23_+_42810055 | 0.15 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr2_+_24867534 | 0.15 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr8_+_7033049 | 0.14 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr17_-_44968177 | 0.14 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr23_-_26536055 | 0.13 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr5_+_38886499 | 0.12 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr22_+_26853254 | 0.12 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr19_-_42928979 | 0.12 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr17_+_23578316 | 0.11 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr5_-_1047504 | 0.10 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr7_+_11390462 | 0.10 |
ENSDART00000114383
|
tlnrd1
|
talin rod domain containing 1 |
| chr7_+_38509333 | 0.09 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr24_-_35707552 | 0.08 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr22_-_18116635 | 0.07 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr17_-_23674495 | 0.07 |
ENSDART00000122209
|
ptena
|
phosphatase and tensin homolog A |
| chr16_-_45854882 | 0.06 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr5_-_1047222 | 0.05 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr1_-_9644630 | 0.04 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr2_+_13721888 | 0.04 |
ENSDART00000089618
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr9_+_54158387 | 0.03 |
ENSDART00000171315
ENSDART00000180570 |
TLR8
|
toll like receptor 8 |
| chr2_+_13722116 | 0.03 |
ENSDART00000155015
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr5_-_22001003 | 0.02 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr24_-_38574631 | 0.02 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr18_+_33100606 | 0.01 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr12_-_145076 | 0.01 |
ENSDART00000160926
|
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr18_+_17827149 | 0.01 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr18_+_17151916 | 0.01 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr23_-_36934944 | 0.00 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr19_-_36640677 | 0.00 |
ENSDART00000189743
|
csmd2
|
CUB and Sushi multiple domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of glis2a+glis2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |