Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
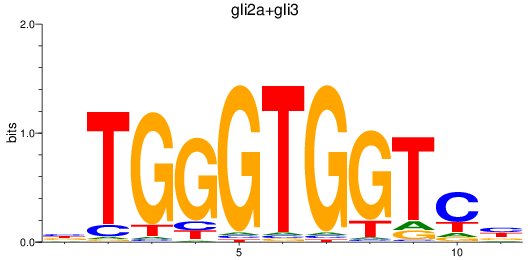
Results for gli2a+gli3
Z-value: 1.11
Transcription factors associated with gli2a+gli3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2a
|
ENSDARG00000025641 | GLI family zinc finger 2a |
|
gli3
|
ENSDARG00000052131 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli3 | dr11_v1_chr24_-_11821505_11821505 | 0.72 | 3.0e-02 | Click! |
| gli2a | dr11_v1_chr9_+_37152564_37152564 | 0.37 | 3.3e-01 | Click! |
Activity profile of gli2a+gli3 motif
Sorted Z-values of gli2a+gli3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_55131289 | 3.49 |
ENSDART00000111585
|
AL935210.1
|
|
| chr3_+_55114097 | 2.70 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr23_+_20422661 | 2.57 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr3_+_55122662 | 2.29 |
ENSDART00000128380
|
hbbe1.2
|
hemoglobin beta embryonic-1.2 |
| chr1_-_19845378 | 2.23 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr7_-_8374950 | 2.17 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr10_-_22854758 | 1.92 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr12_+_27117609 | 1.33 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr5_+_51594209 | 1.23 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr25_-_28384954 | 1.18 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr19_-_26235219 | 1.15 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr12_-_464007 | 1.14 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr22_+_38173960 | 1.13 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr8_-_14484599 | 1.09 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr8_+_31119548 | 1.09 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr10_-_8358396 | 1.08 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr13_+_42124566 | 1.07 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr8_+_29962635 | 1.02 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr8_-_47800754 | 1.01 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr23_+_28582865 | 1.01 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr22_+_38762693 | 1.01 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr14_+_25465346 | 0.96 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr10_+_18952271 | 0.93 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr19_+_56351 | 0.93 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr19_-_10243148 | 0.93 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr11_+_37250839 | 0.92 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr12_+_24562667 | 0.84 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr14_-_215051 | 0.83 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr6_-_35106425 | 0.80 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr18_-_14836600 | 0.77 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr13_+_3252950 | 0.77 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr7_-_15413019 | 0.75 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr12_-_17479078 | 0.75 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr24_+_18299175 | 0.74 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr10_-_35542071 | 0.73 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr25_-_210730 | 0.72 |
ENSDART00000187580
|
FP236318.1
|
|
| chr4_+_5132951 | 0.69 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr13_+_3954540 | 0.68 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr2_-_33993533 | 0.67 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr25_+_7461624 | 0.66 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr4_-_75175407 | 0.66 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr6_-_35446110 | 0.65 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr1_-_470812 | 0.63 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr20_+_15552657 | 0.63 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr16_-_37964325 | 0.62 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr1_+_51479128 | 0.58 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr7_-_15413188 | 0.58 |
ENSDART00000185980
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr25_+_30298377 | 0.57 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr5_-_43959972 | 0.56 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr20_+_26095530 | 0.56 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr22_-_10487490 | 0.55 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr9_-_1984604 | 0.53 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr17_-_42213822 | 0.52 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr21_+_22985078 | 0.52 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr13_-_40726865 | 0.51 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr13_+_30804367 | 0.50 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr9_+_6587364 | 0.50 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr20_+_27194833 | 0.50 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr7_+_73780936 | 0.49 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr16_-_38067952 | 0.47 |
ENSDART00000114266
|
plekho1b
|
pleckstrin homology domain containing, family O member 1b |
| chr14_+_25464681 | 0.45 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr8_-_52859301 | 0.45 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr5_-_23179319 | 0.44 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr10_-_1827225 | 0.43 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_51783120 | 0.43 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr13_-_30150592 | 0.43 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr5_+_20693724 | 0.43 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr9_+_6587056 | 0.43 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr6_-_49655879 | 0.42 |
ENSDART00000191594
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr16_-_31644545 | 0.41 |
ENSDART00000181634
|
CR855311.5
|
|
| chr6_-_10320676 | 0.41 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr5_-_14564878 | 0.40 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr9_-_12736089 | 0.40 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr15_-_35252522 | 0.40 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr6_+_7123980 | 0.40 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr10_-_24868581 | 0.39 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr16_+_37470717 | 0.39 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr8_-_19216657 | 0.38 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr17_+_30587333 | 0.38 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr1_+_7956030 | 0.37 |
ENSDART00000159655
|
CR855320.1
|
|
| chr6_+_54358529 | 0.37 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr21_+_10739846 | 0.35 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr17_+_15388479 | 0.32 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr10_+_26747755 | 0.32 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr21_-_45073764 | 0.31 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr21_-_5856050 | 0.31 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr20_-_28642061 | 0.30 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr20_-_915234 | 0.30 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr25_-_17403598 | 0.29 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr25_+_32473433 | 0.29 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr17_-_51202339 | 0.28 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr20_-_38610360 | 0.28 |
ENSDART00000168783
ENSDART00000164638 |
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr16_+_20220225 | 0.27 |
ENSDART00000182208
|
colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr18_+_22793743 | 0.27 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr14_+_43554490 | 0.27 |
ENSDART00000060351
|
adra1bb
|
adrenoceptor alpha 1Bb |
| chr3_+_57991074 | 0.26 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr25_+_32474031 | 0.25 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr15_+_16525126 | 0.25 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr25_+_245438 | 0.25 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr11_+_30314885 | 0.25 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr10_-_15048781 | 0.25 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr1_-_17797802 | 0.24 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr4_+_4902392 | 0.23 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr14_-_29859067 | 0.19 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr18_-_17485419 | 0.19 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr25_+_22730490 | 0.18 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr7_+_34790768 | 0.18 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr18_+_9641210 | 0.17 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr24_-_25184553 | 0.17 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_-_43822179 | 0.17 |
ENSDART00000185246
|
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr12_-_46145635 | 0.17 |
ENSDART00000074682
|
zgc:153932
|
zgc:153932 |
| chr5_-_58112032 | 0.17 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr24_+_7828613 | 0.15 |
ENSDART00000135161
|
zgc:101569
|
zgc:101569 |
| chr23_-_27875140 | 0.15 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr8_-_49201258 | 0.15 |
ENSDART00000114584
ENSDART00000142172 |
zgc:171501
|
zgc:171501 |
| chr13_+_12396316 | 0.14 |
ENSDART00000143557
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr8_-_40075983 | 0.14 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr24_-_6258869 | 0.14 |
ENSDART00000006636
ENSDART00000178667 |
myo3a
|
myosin IIIA |
| chr20_+_10498704 | 0.13 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr3_+_23692462 | 0.13 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr16_-_4836218 | 0.13 |
ENSDART00000054077
|
tpbgb
|
trophoblast glycoprotein b |
| chr6_-_8480815 | 0.13 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr10_+_40235959 | 0.12 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr21_-_30415524 | 0.12 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr5_+_20437124 | 0.12 |
ENSDART00000159001
|
tmem119a
|
transmembrane protein 119a |
| chr25_-_27722614 | 0.12 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr21_-_30935262 | 0.12 |
ENSDART00000172390
|
stim1b
|
stromal interaction molecule 1b |
| chr23_+_12072980 | 0.11 |
ENSDART00000141646
|
edn3b
|
endothelin 3b |
| chr13_-_45201300 | 0.10 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr21_+_29227224 | 0.10 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr13_+_31583034 | 0.10 |
ENSDART00000111763
|
six6a
|
SIX homeobox 6a |
| chr23_+_21424747 | 0.10 |
ENSDART00000135440
ENSDART00000104214 |
tas1r1
|
taste receptor, type 1, member 1 |
| chr20_+_40457599 | 0.09 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr17_+_6563307 | 0.09 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr4_-_17741513 | 0.09 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr21_-_38619305 | 0.09 |
ENSDART00000139178
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr4_-_38476901 | 0.08 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr3_-_19091024 | 0.08 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr18_+_33100606 | 0.08 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr21_+_27340682 | 0.06 |
ENSDART00000011305
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr2_-_30784198 | 0.06 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr5_-_25621386 | 0.06 |
ENSDART00000051565
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr20_-_25626693 | 0.06 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr8_+_28467893 | 0.06 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr23_+_4414164 | 0.05 |
ENSDART00000192762
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr9_-_11655031 | 0.05 |
ENSDART00000044314
|
itgav
|
integrin, alpha V |
| chr7_+_51816321 | 0.05 |
ENSDART00000073846
|
si:ch211-122f10.4
|
si:ch211-122f10.4 |
| chr22_-_188102 | 0.05 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr23_+_4414343 | 0.04 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr2_+_48638827 | 0.03 |
ENSDART00000163634
|
CR381673.2
|
|
| chr18_+_3022188 | 0.03 |
ENSDART00000170004
|
CU570791.1
|
|
| chr19_-_10631482 | 0.03 |
ENSDART00000129264
|
si:dkey-211g8.9
|
si:dkey-211g8.9 |
| chr3_+_5083407 | 0.03 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr6_-_43817761 | 0.03 |
ENSDART00000183945
|
foxp1b
|
forkhead box P1b |
| chr10_-_36726188 | 0.03 |
ENSDART00000157262
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr14_+_46020282 | 0.02 |
ENSDART00000190087
|
c1qtnf2
|
C1q and TNF related 2 |
| chr18_+_9635178 | 0.02 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr19_+_2546775 | 0.02 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr23_-_14057191 | 0.01 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr20_-_35052464 | 0.01 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr16_-_21047872 | 0.00 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr16_-_4026265 | 0.00 |
ENSDART00000149339
|
si:ch211-175f12.2
|
si:ch211-175f12.2 |
| chr23_-_41824460 | 0.00 |
ENSDART00000131235
|
GMEB2
|
si:ch73-302a13.2 |
| chr18_+_2585298 | 0.00 |
ENSDART00000166314
ENSDART00000162043 |
p2ry2.2
|
purinergic receptor P2Y2, tandem duplicate 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gli2a+gli3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.2 | 0.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.7 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.2 | 1.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 0.5 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 2.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.1 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.6 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.4 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 0.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.7 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 2.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.4 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 2.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 2.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 2.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 1.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.7 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.7 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 2.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.5 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.2 | 0.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 0.6 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |