Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for gfi1b
Z-value: 0.74
Transcription factors associated with gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
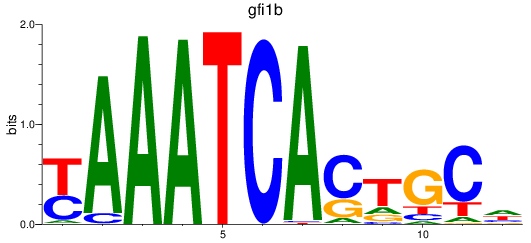
gfi1b
|
ENSDARG00000079947 | growth factor independent 1B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1b | dr11_v1_chr21_-_17296010_17296010 | 0.52 | 1.5e-01 | Click! |
Activity profile of gfi1b motif
Sorted Z-values of gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433518 | 1.84 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 1.73 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr3_-_32818607 | 1.57 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_-_45235947 | 1.45 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr23_-_35694171 | 1.30 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr5_-_40734045 | 1.23 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr23_-_35694461 | 1.22 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr7_+_48319916 | 1.02 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr24_-_32408404 | 1.00 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr17_-_12385308 | 0.94 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_-_24103666 | 0.90 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr25_+_19999623 | 0.89 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr1_-_25679339 | 0.85 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr6_+_29791164 | 0.81 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr9_+_48007081 | 0.76 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr25_-_15049694 | 0.74 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr9_+_34641237 | 0.68 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr20_+_28434196 | 0.67 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr23_-_24263474 | 0.65 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr17_-_21066075 | 0.64 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr20_-_26042070 | 0.64 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr5_-_36328688 | 0.64 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr9_+_30108641 | 0.62 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr3_-_12187245 | 0.61 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr21_-_4032650 | 0.61 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr13_+_13681681 | 0.57 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr3_-_32541033 | 0.56 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr19_+_42609132 | 0.52 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr17_-_12389259 | 0.52 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_+_35362225 | 0.51 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr23_+_22656477 | 0.51 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr7_-_26408472 | 0.50 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr3_-_26109322 | 0.49 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr7_+_44445595 | 0.46 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr1_+_12763461 | 0.46 |
ENSDART00000159226
ENSDART00000180121 |
pcdh10a
|
protocadherin 10a |
| chr9_-_1965727 | 0.46 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr3_-_30941362 | 0.46 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr7_-_35083184 | 0.46 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr12_+_6002715 | 0.45 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr16_+_10422836 | 0.42 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr21_-_131236 | 0.41 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr13_+_21779975 | 0.40 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr5_-_40510397 | 0.40 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr16_+_23282655 | 0.39 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr10_-_39153959 | 0.39 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr23_-_9768700 | 0.38 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr3_-_12970418 | 0.38 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr11_+_35050253 | 0.37 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr13_-_40398556 | 0.36 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr16_+_43152727 | 0.35 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr3_+_23691847 | 0.35 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr20_+_50852356 | 0.34 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr14_-_21218891 | 0.34 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr7_-_25895189 | 0.33 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr14_+_35023923 | 0.32 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr10_+_8968203 | 0.32 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr22_+_16446052 | 0.32 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr21_+_9628854 | 0.31 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr19_+_47405867 | 0.31 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr2_-_10338759 | 0.31 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr19_+_4912817 | 0.31 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_+_25920792 | 0.31 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr4_-_12007404 | 0.30 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr24_-_4450238 | 0.30 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr19_+_29798064 | 0.29 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr25_+_8955530 | 0.29 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr1_+_1805294 | 0.28 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr5_-_12093618 | 0.28 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr18_+_13203831 | 0.28 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr2_-_30668580 | 0.28 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr2_+_9821757 | 0.27 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr11_+_25634041 | 0.26 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr5_-_23362602 | 0.26 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr12_-_4220713 | 0.26 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr13_-_43149063 | 0.26 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr18_-_16181952 | 0.25 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_-_322769 | 0.25 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr21_-_33995213 | 0.25 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr24_+_35387517 | 0.25 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr6_+_7466223 | 0.25 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr5_+_9259971 | 0.25 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr24_+_9412450 | 0.24 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr15_+_45643787 | 0.24 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr2_+_9822319 | 0.24 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr6_-_10780698 | 0.24 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr4_-_42242844 | 0.24 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr5_+_9259574 | 0.24 |
ENSDART00000185560
|
susd1
|
sushi domain containing 1 |
| chr2_+_54482603 | 0.23 |
ENSDART00000130977
ENSDART00000183090 |
mtcl1
|
microtubule crosslinking factor 1 |
| chr8_-_19395110 | 0.23 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr21_-_17603182 | 0.23 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr15_-_41290415 | 0.23 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr6_-_32349153 | 0.23 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr10_+_9159279 | 0.22 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr11_-_6188413 | 0.22 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr13_-_47403154 | 0.22 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr4_+_28356606 | 0.22 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr4_-_63923523 | 0.22 |
ENSDART00000144330
|
si:dkey-179k24.1
|
si:dkey-179k24.1 |
| chr5_+_4575800 | 0.22 |
ENSDART00000180952
|
cst14b.2
|
cystatin 14b, tandem duplicate 2 |
| chr7_-_28148310 | 0.21 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr22_-_26175237 | 0.21 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr1_+_9860381 | 0.19 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr3_+_22377312 | 0.19 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr8_-_46525092 | 0.18 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr6_-_12851888 | 0.18 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr8_-_52715911 | 0.18 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr17_+_11675362 | 0.18 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr11_-_15296805 | 0.18 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr20_+_18163355 | 0.18 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr6_+_11249706 | 0.18 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr24_-_23716097 | 0.18 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr14_-_49286773 | 0.17 |
ENSDART00000084104
|
n4bp3
|
NEDD4 binding protein 3 |
| chr19_-_28130658 | 0.17 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr14_-_47314340 | 0.17 |
ENSDART00000164851
|
fstl5
|
follistatin-like 5 |
| chr25_-_518656 | 0.17 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr3_-_39695856 | 0.17 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr9_+_19489514 | 0.17 |
ENSDART00000152032
ENSDART00000114256 ENSDART00000190572 ENSDART00000147571 ENSDART00000151918 ENSDART00000152034 |
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr21_+_16145980 | 0.16 |
ENSDART00000135119
|
qrfpr4
|
pyroglutamylated RFamide peptide receptor 4 |
| chr6_-_34220641 | 0.16 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr19_+_21919856 | 0.16 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr10_-_34871737 | 0.16 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr1_+_14466860 | 0.15 |
ENSDART00000114097
|
stim2a
|
tromal interaction molecule 2a |
| chr23_+_25232411 | 0.15 |
ENSDART00000138974
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr7_-_35036770 | 0.15 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr17_+_24687338 | 0.15 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr6_+_3516695 | 0.14 |
ENSDART00000109576
|
faah
|
fatty acid amide hydrolase |
| chr14_-_11529311 | 0.14 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr7_-_24005268 | 0.13 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr6_+_11250033 | 0.13 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_55277419 | 0.13 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr17_+_34805897 | 0.12 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr6_-_23931442 | 0.12 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr25_-_3979583 | 0.12 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr4_-_13567387 | 0.12 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr12_+_48241841 | 0.12 |
ENSDART00000168616
|
ppa1a
|
pyrophosphatase (inorganic) 1a |
| chr21_-_2935455 | 0.11 |
ENSDART00000163963
|
CABZ01023255.1
|
|
| chr14_+_3944826 | 0.11 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr9_+_37152564 | 0.11 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr9_+_21277846 | 0.11 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr3_-_39696066 | 0.10 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr20_+_18163821 | 0.09 |
ENSDART00000186507
|
aqp4
|
aquaporin 4 |
| chr2_-_36925561 | 0.09 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr19_-_22488952 | 0.08 |
ENSDART00000179856
ENSDART00000141503 |
pleca
|
plectin a |
| chr25_+_19041329 | 0.08 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr5_-_62306819 | 0.08 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr17_-_33552363 | 0.08 |
ENSDART00000154400
|
tmem121aa
|
transmembrane protein 121Aa |
| chr14_-_47391084 | 0.08 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr1_-_59139599 | 0.08 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr20_+_27464721 | 0.08 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr13_-_42724645 | 0.08 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr23_+_7471072 | 0.08 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr2_-_25120197 | 0.07 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr21_+_39963851 | 0.07 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr12_+_8474868 | 0.06 |
ENSDART00000062858
|
adoa
|
2-aminoethanethiol (cysteamine) dioxygenase a |
| chr23_-_10137322 | 0.06 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr19_-_35229336 | 0.06 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr17_-_37395460 | 0.06 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr7_+_39360797 | 0.06 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr22_+_31821815 | 0.06 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr11_-_15297129 | 0.06 |
ENSDART00000167985
ENSDART00000163031 ENSDART00000165010 ENSDART00000171250 ENSDART00000010684 ENSDART00000191714 ENSDART00000191164 |
rpn2
|
ribophorin II |
| chr23_+_25232711 | 0.06 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr16_-_263658 | 0.05 |
ENSDART00000129303
|
slc6a3
|
solute carrier family 6 (neurotransmitter transporter), member 3 |
| chr5_+_45976130 | 0.05 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr9_+_19489304 | 0.05 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr25_-_16818978 | 0.04 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr8_+_37111643 | 0.04 |
ENSDART00000061336
|
ren
|
renin |
| chr5_+_51909740 | 0.04 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr11_+_25044082 | 0.03 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr17_+_49127003 | 0.03 |
ENSDART00000157360
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr12_+_4845448 | 0.03 |
ENSDART00000160666
|
ghrhr2
|
growth hormone releasing hormone receptor 2 |
| chr7_+_66739166 | 0.03 |
ENSDART00000154634
|
mrvi1
|
murine retrovirus integration site 1 homolog |
| chr5_-_36924509 | 0.03 |
ENSDART00000025013
|
kptn
|
kaptin (actin binding protein) |
| chr6_+_3516926 | 0.02 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
| chr2_-_24505185 | 0.02 |
ENSDART00000052065
|
si:rp71-39b20.4
|
si:rp71-39b20.4 |
| chr8_-_15182232 | 0.02 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr9_-_459910 | 0.02 |
ENSDART00000162551
|
si:dkey-11f4.16
|
si:dkey-11f4.16 |
| chr21_-_16219400 | 0.02 |
ENSDART00000124871
|
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr14_-_45512807 | 0.02 |
ENSDART00000173172
|
si:ch211-114c17.1
|
si:ch211-114c17.1 |
| chr23_+_23232136 | 0.01 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr20_+_34596205 | 0.01 |
ENSDART00000138338
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr10_-_34867401 | 0.01 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_45728620 | 0.00 |
ENSDART00000188795
|
BX088696.1
|
|
| chr5_-_58832332 | 0.00 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr12_+_38563073 | 0.00 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 1.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.7 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.7 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.3 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.2 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.2 | GO:0009298 | mannose metabolic process(GO:0006013) GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.0 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.0 | 0.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.0 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.5 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.3 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.5 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005334 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |