Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
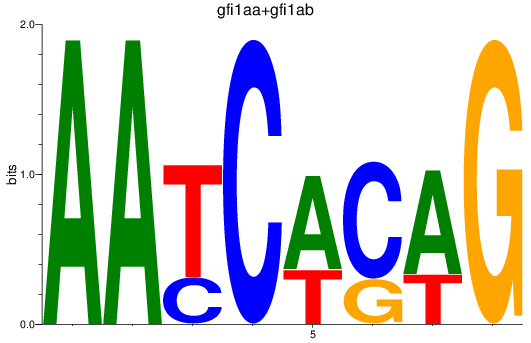
Results for gfi1aa+gfi1ab
Z-value: 1.08
Transcription factors associated with gfi1aa+gfi1ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1aa
|
ENSDARG00000020746 | growth factor independent 1A transcription repressor a |
|
gfi1ab
|
ENSDARG00000044457 | growth factor independent 1A transcription repressor b |
|
gfi1ab
|
ENSDARG00000114140 | growth factor independent 1A transcription repressor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1ab | dr11_v1_chr6_-_29007493_29007493 | 0.90 | 8.7e-04 | Click! |
| gfi1aa | dr11_v1_chr2_+_10766744_10766744 | 0.67 | 4.9e-02 | Click! |
Activity profile of gfi1aa+gfi1ab motif
Sorted Z-values of gfi1aa+gfi1ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433518 | 2.87 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 2.71 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr13_+_4671698 | 2.45 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr19_-_10551394 | 2.02 |
ENSDART00000186815
ENSDART00000181041 |
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr3_-_32818607 | 1.76 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr9_-_18877597 | 1.65 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr16_+_20915319 | 1.61 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr12_+_27127139 | 1.47 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr21_+_26697536 | 1.47 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr4_-_16330368 | 1.29 |
ENSDART00000128932
|
epyc
|
epiphycan |
| chr7_+_38750871 | 1.27 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr13_-_39159810 | 1.15 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr23_+_44883805 | 1.13 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr9_+_48007081 | 1.13 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr9_-_3400727 | 1.12 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr1_-_38195012 | 1.06 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr17_-_21066075 | 1.01 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr13_-_39160018 | 1.00 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr8_+_15254564 | 0.99 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr12_+_25600685 | 0.98 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr5_-_19400166 | 0.97 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr20_+_23238833 | 0.97 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr17_-_36936649 | 0.96 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr17_-_36936856 | 0.95 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr6_+_29217392 | 0.94 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr23_-_21453614 | 0.94 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr18_-_1185772 | 0.94 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr15_-_24869826 | 0.94 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr13_-_46429220 | 0.93 |
ENSDART00000149125
ENSDART00000098269 ENSDART00000150061 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr10_+_39084354 | 0.90 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr12_+_22870372 | 0.90 |
ENSDART00000182264
ENSDART00000191789 ENSDART00000130594 ENSDART00000190433 |
afap1
|
actin filament associated protein 1 |
| chr6_+_35362225 | 0.90 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr21_+_35215810 | 0.89 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr2_-_16562505 | 0.88 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr23_-_21471022 | 0.88 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr14_-_40389699 | 0.87 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr17_+_12698532 | 0.87 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr11_+_37201483 | 0.86 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr25_-_15049694 | 0.86 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr20_-_48485354 | 0.86 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr16_-_20312146 | 0.85 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr9_-_45602978 | 0.85 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr17_-_6382392 | 0.84 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr16_+_29650698 | 0.84 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr23_+_21459263 | 0.82 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr1_-_20271138 | 0.82 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_+_54013457 | 0.82 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr6_-_13783604 | 0.82 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr15_+_19652807 | 0.82 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr14_+_33722950 | 0.82 |
ENSDART00000075312
|
apln
|
apelin |
| chr14_+_35806605 | 0.81 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr5_+_64732270 | 0.81 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr5_+_43603794 | 0.80 |
ENSDART00000134633
|
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr15_+_36115955 | 0.80 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr17_+_26569601 | 0.80 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr23_+_8797143 | 0.79 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr25_+_21829777 | 0.78 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_+_26629084 | 0.78 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr15_+_45643787 | 0.78 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr12_-_31103187 | 0.78 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr16_+_23397785 | 0.78 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr3_+_23703704 | 0.76 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr7_-_51546386 | 0.76 |
ENSDART00000174306
|
nhsl2
|
NHS-like 2 |
| chr22_+_16022211 | 0.75 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr23_+_21455152 | 0.75 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr11_+_30310170 | 0.75 |
ENSDART00000127797
|
ugt1b3
|
UDP glucuronosyltransferase 1 family, polypeptide B3 |
| chr13_+_25433774 | 0.74 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr3_+_31621774 | 0.74 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr9_+_3388099 | 0.73 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr4_+_6643421 | 0.73 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr21_+_20771082 | 0.73 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr5_-_66702479 | 0.72 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr4_-_16412084 | 0.72 |
ENSDART00000188460
|
dcn
|
decorin |
| chr16_+_11724230 | 0.72 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr18_-_16123222 | 0.71 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr12_+_22258962 | 0.71 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr10_+_16592851 | 0.70 |
ENSDART00000187508
ENSDART00000101142 |
chsy3
|
chondroitin sulfate synthase 3 |
| chr25_-_23526058 | 0.70 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr23_+_22656477 | 0.69 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr5_+_70155935 | 0.68 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr5_-_65782783 | 0.68 |
ENSDART00000130888
ENSDART00000050855 |
notch1b
|
notch 1b |
| chr21_-_26495700 | 0.67 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr21_-_44104600 | 0.66 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr7_-_26408472 | 0.66 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr23_+_36101185 | 0.66 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr21_-_4032650 | 0.66 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr1_-_54947592 | 0.65 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr11_+_21910343 | 0.64 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr11_+_21910752 | 0.64 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr18_+_16744307 | 0.64 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr7_-_24364536 | 0.63 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr11_-_39044595 | 0.63 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr14_-_17563773 | 0.63 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr21_-_36972127 | 0.63 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr16_+_23282655 | 0.63 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr13_-_22862133 | 0.62 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr5_+_17727310 | 0.62 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr1_-_46981134 | 0.62 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr6_+_1787160 | 0.62 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr21_+_30351256 | 0.62 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr23_+_36095260 | 0.61 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr8_+_37700090 | 0.61 |
ENSDART00000187885
ENSDART00000127633 |
adrb3a
|
adrenoceptor beta 3a |
| chr24_+_14713776 | 0.61 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr15_-_19250543 | 0.61 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr22_-_13042992 | 0.61 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr10_+_35265083 | 0.61 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr13_-_16222388 | 0.60 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr8_+_39619087 | 0.60 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr21_-_10773344 | 0.60 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr11_+_30306606 | 0.60 |
ENSDART00000128276
ENSDART00000190222 |
ugt1b4
|
UDP glucuronosyltransferase 1 family, polypeptide B4 |
| chr13_+_27951688 | 0.59 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr17_-_32863250 | 0.59 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr7_-_60831082 | 0.59 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr7_+_40638210 | 0.59 |
ENSDART00000052236
|
mnx1
|
motor neuron and pancreas homeobox 1 |
| chr25_+_14017609 | 0.59 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr13_+_51579851 | 0.59 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr12_+_6002715 | 0.59 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr19_+_4912817 | 0.58 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr4_+_18960914 | 0.58 |
ENSDART00000139730
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr21_-_43131752 | 0.58 |
ENSDART00000024137
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr24_-_8729531 | 0.58 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr12_+_2399130 | 0.57 |
ENSDART00000075179
|
vstm4b
|
V-set and transmembrane domain containing 4b |
| chr15_+_22311803 | 0.57 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr14_+_7452302 | 0.57 |
ENSDART00000130388
|
gfra3
|
GDNF family receptor alpha 3 |
| chr18_+_38749547 | 0.56 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr1_+_59538755 | 0.56 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr10_+_43994471 | 0.56 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr5_-_36948586 | 0.56 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr7_+_56098590 | 0.56 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr16_+_813780 | 0.56 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr8_+_54055390 | 0.56 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr8_-_19216657 | 0.56 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr3_+_19621034 | 0.55 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr19_-_17774875 | 0.55 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr9_-_7684002 | 0.55 |
ENSDART00000016360
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr9_-_7683799 | 0.55 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr23_+_36178104 | 0.55 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr1_+_54037077 | 0.55 |
ENSDART00000109386
|
triobpa
|
TRIO and F-actin binding protein a |
| chr11_+_25634041 | 0.55 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr5_-_12219572 | 0.55 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr5_-_28029558 | 0.55 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr16_-_42894628 | 0.55 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr8_+_1766206 | 0.54 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr15_-_21014015 | 0.54 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr4_-_7212875 | 0.52 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr18_+_17583479 | 0.52 |
ENSDART00000186977
ENSDART00000010998 |
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr6_+_42819337 | 0.52 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr6_-_19023468 | 0.51 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr17_-_26911852 | 0.51 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr3_+_40170216 | 0.51 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr2_-_32558795 | 0.51 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr6_-_48094342 | 0.50 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr17_-_12385308 | 0.50 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr20_-_31427390 | 0.50 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr9_-_1965727 | 0.50 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr9_-_18716 | 0.50 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr10_+_21722892 | 0.50 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr21_-_23475361 | 0.50 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr25_+_13406069 | 0.50 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr1_-_51734524 | 0.49 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr13_+_21779975 | 0.49 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr14_+_34514336 | 0.49 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr7_+_32369026 | 0.48 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr14_-_40390757 | 0.48 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr5_-_36328688 | 0.48 |
ENSDART00000011399
|
efnb1
|
ephrin-B1 |
| chr20_-_26491567 | 0.48 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr4_-_2545310 | 0.48 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr16_-_29528198 | 0.48 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr14_-_41678357 | 0.48 |
ENSDART00000185925
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr14_-_39074539 | 0.48 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr4_+_4803698 | 0.47 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr7_-_22132265 | 0.47 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr10_+_29265463 | 0.47 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr5_+_38276582 | 0.47 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr20_-_20533865 | 0.46 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr24_+_4977862 | 0.45 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr7_-_12968689 | 0.45 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr8_+_26192930 | 0.45 |
ENSDART00000137391
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr20_+_52554352 | 0.45 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr8_+_29962635 | 0.44 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr18_-_14836600 | 0.44 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr9_+_23255410 | 0.44 |
ENSDART00000113241
ENSDART00000137231 |
tmem163a
|
transmembrane protein 163a |
| chr8_-_14179798 | 0.44 |
ENSDART00000040645
ENSDART00000146749 |
rhoaa
|
ras homolog gene family, member Aa |
| chr14_+_49135264 | 0.44 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr14_+_30491890 | 0.44 |
ENSDART00000131174
|
fgf20b
|
fibroblast growth factor 20b |
| chr21_-_14310159 | 0.44 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
| chr13_-_7575216 | 0.44 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr2_+_48288461 | 0.43 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr23_+_22200467 | 0.43 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr2_-_10062575 | 0.43 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr25_-_5963535 | 0.43 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr8_+_24747865 | 0.43 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr8_-_2434282 | 0.42 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr18_+_5490668 | 0.42 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr9_-_1984604 | 0.42 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr4_+_22480169 | 0.42 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr14_+_33723309 | 0.42 |
ENSDART00000132488
|
apln
|
apelin |
| chr7_+_25920792 | 0.42 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr15_+_28368644 | 0.42 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr24_-_26304386 | 0.42 |
ENSDART00000175416
|
otos
|
otospiralin |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1aa+gfi1ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 1.0 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.3 | 1.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 1.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 2.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 0.8 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.2 | 0.7 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 1.7 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 1.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.3 | GO:0006953 | acute inflammatory response(GO:0002526) acute-phase response(GO:0006953) |
| 0.2 | 1.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 4.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 1.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 0.8 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.2 | 0.5 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.2 | 2.5 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.5 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.7 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.9 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 0.8 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.9 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.6 | GO:0048909 | anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.3 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 1.5 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 1.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.8 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.8 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.6 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.3 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.3 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.8 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.7 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 1.8 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.5 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.9 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.5 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.3 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) |
| 0.0 | 0.5 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.8 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.6 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.5 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:2001057 | reactive nitrogen species metabolic process(GO:2001057) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.2 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0050891 | respiratory burst(GO:0045730) multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 1.4 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.4 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.3 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.7 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0097028 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.3 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:1904949 | ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.7 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.4 | 5.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 0.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 0.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.2 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.4 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 2.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.7 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.0 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0098918 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 19.3 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |