Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
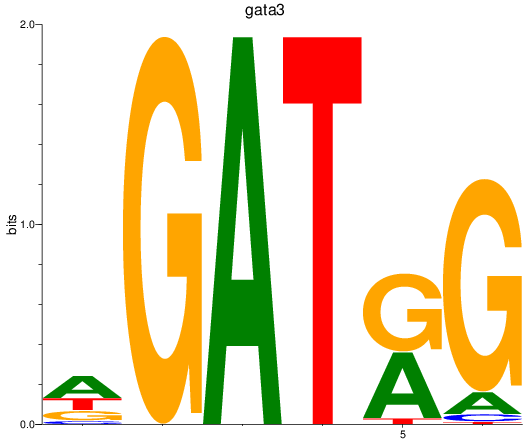
Results for gata3
Z-value: 1.08
Transcription factors associated with gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata3
|
ENSDARG00000016526 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata3 | dr11_v1_chr4_-_25064510_25064510 | -0.26 | 5.0e-01 | Click! |
Activity profile of gata3 motif
Sorted Z-values of gata3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13382852 | 2.27 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr16_-_35532937 | 2.16 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr11_-_40457325 | 2.01 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr8_-_6825588 | 1.28 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr9_-_35557397 | 1.21 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr4_-_16628801 | 1.20 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr23_-_41762956 | 1.18 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr22_-_3299355 | 1.16 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr15_+_31735931 | 1.01 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr5_+_8964926 | 0.99 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr6_+_2174082 | 0.96 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr22_+_21398508 | 0.91 |
ENSDART00000089408
ENSDART00000186091 |
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr5_+_44654535 | 0.84 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr23_+_35426404 | 0.81 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr21_-_3844322 | 0.80 |
ENSDART00000166652
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr5_-_3960161 | 0.77 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr2_-_42705284 | 0.76 |
ENSDART00000187160
|
myo10
|
myosin X |
| chr1_-_14258409 | 0.76 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr12_+_48581588 | 0.75 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr5_+_44655148 | 0.73 |
ENSDART00000124059
|
dapk1
|
death-associated protein kinase 1 |
| chr25_+_1335530 | 0.71 |
ENSDART00000090803
|
fem1b
|
fem-1 homolog b (C. elegans) |
| chr10_-_31175744 | 0.69 |
ENSDART00000191728
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr23_-_41762797 | 0.66 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr23_-_28423222 | 0.65 |
ENSDART00000182058
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr20_-_34754617 | 0.63 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr17_-_5732299 | 0.62 |
ENSDART00000017244
|
xkr6a
|
XK, Kell blood group complex subunit-related family, member 6a |
| chr21_-_7940043 | 0.61 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr3_-_40768548 | 0.59 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr21_-_41369539 | 0.59 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr3_+_51684963 | 0.57 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr12_+_38770654 | 0.57 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr22_+_21549419 | 0.55 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr18_+_35229115 | 0.55 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr1_+_1941031 | 0.54 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr16_-_16649764 | 0.54 |
ENSDART00000014439
|
tnxba
|
tenascin XBa |
| chr1_-_33645967 | 0.49 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr10_+_37182626 | 0.49 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr20_+_22130284 | 0.49 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr10_-_2527342 | 0.49 |
ENSDART00000184168
|
CU856539.1
|
|
| chr25_+_33063762 | 0.48 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr7_-_48173440 | 0.46 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr8_-_22698651 | 0.46 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr24_-_2381143 | 0.46 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr18_+_44769211 | 0.45 |
ENSDART00000177181
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr13_+_20524921 | 0.45 |
ENSDART00000081385
|
ccdc172
|
coiled-coil domain containing 172 |
| chr22_-_3299100 | 0.42 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr20_+_13781779 | 0.41 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr25_+_11389220 | 0.41 |
ENSDART00000187787
|
AGBL1
|
si:ch73-141f14.1 |
| chr11_+_13424116 | 0.39 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr1_-_55238610 | 0.39 |
ENSDART00000110818
|
FO704915.1
|
|
| chr15_+_30126971 | 0.39 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr9_+_44304980 | 0.39 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr18_-_25051846 | 0.39 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr17_+_23554932 | 0.39 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr15_-_9294929 | 0.39 |
ENSDART00000155717
|
si:ch211-261a10.5
|
si:ch211-261a10.5 |
| chr20_-_48061351 | 0.38 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr10_-_10018120 | 0.37 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr7_-_13884610 | 0.37 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr9_+_24192370 | 0.37 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr24_-_29822913 | 0.37 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr20_+_35445462 | 0.36 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr11_-_36020005 | 0.36 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr6_+_37623693 | 0.36 |
ENSDART00000144812
ENSDART00000182709 |
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr19_+_19989380 | 0.35 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr18_-_22753637 | 0.34 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr10_+_9595575 | 0.34 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr22_-_16270462 | 0.33 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr13_+_31648271 | 0.32 |
ENSDART00000006648
|
mnat1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr8_-_8446668 | 0.32 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr15_-_34866219 | 0.31 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr2_+_42872661 | 0.31 |
ENSDART00000036979
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr20_+_35085224 | 0.31 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr1_+_46493944 | 0.31 |
ENSDART00000114083
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr6_+_45346808 | 0.30 |
ENSDART00000186327
|
LO017951.1
|
|
| chr6_-_49159207 | 0.30 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr3_-_13146631 | 0.30 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_-_5624361 | 0.30 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr20_-_51559419 | 0.29 |
ENSDART00000065231
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr12_+_19958845 | 0.29 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr22_+_997838 | 0.29 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr21_-_41369370 | 0.28 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr15_-_16704417 | 0.28 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr9_-_1639884 | 0.28 |
ENSDART00000062850
ENSDART00000150947 |
agps
|
alkylglycerone phosphate synthase |
| chr20_-_15922986 | 0.28 |
ENSDART00000189421
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr25_+_32496723 | 0.28 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr10_+_37181780 | 0.28 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr18_-_20822045 | 0.28 |
ENSDART00000100743
|
cttnbp2
|
cortactin binding protein 2 |
| chr21_+_33249478 | 0.28 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr14_-_35892767 | 0.28 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr17_+_22587356 | 0.28 |
ENSDART00000157328
|
birc6
|
baculoviral IAP repeat containing 6 |
| chr4_+_4889257 | 0.27 |
ENSDART00000175651
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr25_+_32496877 | 0.26 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr7_+_71955486 | 0.26 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr23_+_34321237 | 0.26 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr9_+_45605410 | 0.26 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr16_-_52736549 | 0.25 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
| chr8_-_31417139 | 0.25 |
ENSDART00000180204
|
znf131
|
zinc finger protein 131 |
| chr4_-_19883985 | 0.25 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr7_+_34296789 | 0.25 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_-_874888 | 0.25 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr3_+_15394428 | 0.24 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr17_-_14523722 | 0.24 |
ENSDART00000024726
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr8_+_11325310 | 0.24 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr18_+_20560616 | 0.24 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr19_+_7552699 | 0.24 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr17_+_49500820 | 0.24 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr2_+_54755172 | 0.24 |
ENSDART00000097864
|
ankrd12
|
ankyrin repeat domain 12 |
| chr19_-_977849 | 0.24 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr8_-_33154677 | 0.23 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr14_+_44545092 | 0.23 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr10_-_10018794 | 0.23 |
ENSDART00000130734
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr6_-_40446536 | 0.23 |
ENSDART00000153466
|
tatdn2
|
TatD DNase domain containing 2 |
| chr4_-_21466825 | 0.23 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr10_+_44581378 | 0.23 |
ENSDART00000190331
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr17_-_26604549 | 0.23 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr8_+_12951155 | 0.23 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr20_-_52939501 | 0.22 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_+_51796441 | 0.22 |
ENSDART00000165151
|
crygn1
|
crystallin, gamma N1 |
| chr23_-_20010579 | 0.22 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr20_+_40739964 | 0.22 |
ENSDART00000061251
ENSDART00000156669 |
cx28.1
|
connexin 28.1 |
| chr11_+_13423776 | 0.22 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr16_+_31511739 | 0.22 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr2_+_29997043 | 0.22 |
ENSDART00000139566
ENSDART00000151848 |
rbm33b
|
RNA binding motif protein 33b |
| chr8_+_26859639 | 0.22 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr3_+_23247325 | 0.22 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr5_-_18513950 | 0.22 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr1_-_21321482 | 0.22 |
ENSDART00000054440
|
tmem144a
|
transmembrane protein 144a |
| chr8_+_9699111 | 0.22 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr21_-_34261677 | 0.22 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr12_-_23320266 | 0.21 |
ENSDART00000181711
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr14_+_20156477 | 0.21 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr24_-_26885897 | 0.21 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr4_-_19884440 | 0.21 |
ENSDART00000182979
ENSDART00000105967 |
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr15_-_33304133 | 0.21 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr16_+_48714048 | 0.20 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr18_+_14307059 | 0.20 |
ENSDART00000186558
|
zgc:173742
|
zgc:173742 |
| chr12_+_28986308 | 0.20 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr9_+_50007556 | 0.20 |
ENSDART00000175587
|
slc38a11
|
solute carrier family 38, member 11 |
| chr18_+_3169579 | 0.20 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr20_-_165818 | 0.20 |
ENSDART00000123860
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr6_-_31827597 | 0.20 |
ENSDART00000159400
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr2_+_37207461 | 0.20 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr16_-_5873866 | 0.20 |
ENSDART00000136936
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr21_-_41588129 | 0.19 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr22_+_15633013 | 0.19 |
ENSDART00000188095
ENSDART00000048763 |
CABZ01090041.1
|
|
| chr4_-_72638972 | 0.19 |
ENSDART00000193312
|
CABZ01054394.1
|
|
| chr7_+_17816006 | 0.19 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr11_+_36989696 | 0.18 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr8_-_410199 | 0.18 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr8_-_49701904 | 0.18 |
ENSDART00000142115
|
frmd3
|
FERM domain containing 3 |
| chr2_-_38992304 | 0.18 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr19_-_36675023 | 0.18 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr16_-_26132122 | 0.18 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr20_+_6535438 | 0.18 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr15_+_44201056 | 0.18 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr13_-_46991577 | 0.18 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr11_-_42230491 | 0.18 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr23_+_2542158 | 0.17 |
ENSDART00000182197
|
LO017835.1
|
|
| chr3_-_21137362 | 0.17 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr17_+_6535694 | 0.17 |
ENSDART00000191681
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr6_-_15604157 | 0.17 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr7_+_72882446 | 0.17 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr6_-_6295513 | 0.17 |
ENSDART00000092172
|
mtif2
|
mitochondrial translational initiation factor 2 |
| chr11_-_13152215 | 0.17 |
ENSDART00000160989
ENSDART00000158239 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr3_-_2623176 | 0.17 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr11_-_2270069 | 0.17 |
ENSDART00000189005
|
znf740a
|
zinc finger protein 740a |
| chr10_-_9192450 | 0.17 |
ENSDART00000139783
|
si:dkeyp-41f9.4
|
si:dkeyp-41f9.4 |
| chr4_-_14315855 | 0.17 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr4_-_20135919 | 0.17 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr10_+_1849874 | 0.17 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr18_+_44769027 | 0.17 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr13_+_33462232 | 0.17 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr3_-_34717882 | 0.17 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr17_-_17447899 | 0.16 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr10_-_35186310 | 0.16 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr19_+_19988869 | 0.16 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr16_-_33097398 | 0.16 |
ENSDART00000166617
|
dopey1
|
dopey family member 1 |
| chr21_-_7781555 | 0.16 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr3_+_53352018 | 0.16 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr8_+_41048501 | 0.16 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr23_-_32334208 | 0.16 |
ENSDART00000053472
|
rnf41
|
ring finger protein 41 |
| chr1_-_53988017 | 0.16 |
ENSDART00000003097
|
RHOU (1 of many)
|
si:ch211-133l11.10 |
| chr8_-_3716734 | 0.16 |
ENSDART00000172966
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr2_-_41964402 | 0.16 |
ENSDART00000131278
|
si:dkey-97a13.6
|
si:dkey-97a13.6 |
| chr8_-_410728 | 0.16 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr6_+_37754763 | 0.16 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr16_+_35535171 | 0.16 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr15_+_29025090 | 0.16 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr9_-_51370293 | 0.16 |
ENSDART00000084806
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr23_-_18415872 | 0.16 |
ENSDART00000135430
|
fam120c
|
family with sequence similarity 120C |
| chr6_+_30491129 | 0.16 |
ENSDART00000088142
|
wwc3
|
WWC family member 3 |
| chr15_-_25099679 | 0.16 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr12_-_22238004 | 0.16 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr19_-_38419575 | 0.15 |
ENSDART00000087639
|
smap2
|
small ArfGAP2 |
| chr24_-_7587401 | 0.15 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr20_-_164300 | 0.15 |
ENSDART00000183354
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr14_+_44805649 | 0.15 |
ENSDART00000180361
|
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr8_+_19621511 | 0.15 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr8_-_8446430 | 0.15 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr5_+_13521081 | 0.15 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.5 | 2.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 1.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.3 | 1.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.2 | 0.6 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 2.0 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.2 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.2 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.1 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.0 | 0.2 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.0 | 0.3 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 1.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.2 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.4 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.3 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 1.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.5 | GO:0097178 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.3 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 1.0 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0060213 | positive regulation of mRNA 3'-end processing(GO:0031442) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.0 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.9 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.1 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:0090133 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.0 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.0 | 0.0 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 0.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 2.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 0.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 1.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.1 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.0 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |