Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
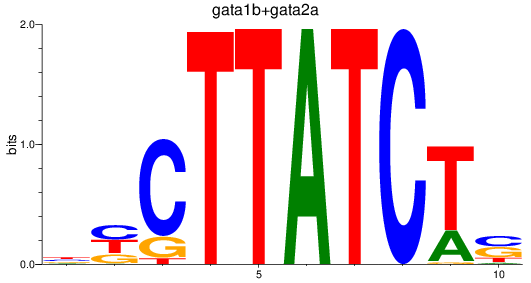
Results for gata1b+gata2a
Z-value: 3.40
Transcription factors associated with gata1b+gata2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata1b
|
ENSDARG00000059130 | GATA binding protein 1b |
|
gata2a
|
ENSDARG00000059327 | GATA binding protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata1b | dr11_v1_chr8_-_7474997_7474997 | 0.76 | 1.6e-02 | Click! |
| gata2a | dr11_v1_chr11_-_3865472_3865472 | 0.51 | 1.6e-01 | Click! |
Activity profile of gata1b+gata2a motif
Sorted Z-values of gata1b+gata2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_20350629 | 7.96 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr14_-_25949713 | 7.83 |
ENSDART00000181455
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr23_-_23401305 | 7.62 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr25_+_21829777 | 7.44 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr14_-_25949951 | 6.70 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr19_+_7567763 | 6.21 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr1_+_25848231 | 5.88 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr1_-_45636362 | 5.87 |
ENSDART00000181757
|
sept3
|
septin 3 |
| chr7_-_51102479 | 5.85 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr20_+_15552657 | 5.75 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr13_+_2908764 | 5.67 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr5_-_64168415 | 5.57 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr5_+_15203421 | 5.52 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr10_+_10801564 | 5.45 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr16_+_51180938 | 5.16 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_+_10801719 | 5.03 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr7_-_73752955 | 4.99 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr8_+_16004551 | 4.91 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_-_50287949 | 4.75 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr22_+_16535575 | 4.74 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr21_-_11834452 | 4.59 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr3_-_35602233 | 4.48 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr15_-_21877726 | 4.44 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr4_+_1530287 | 4.31 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr9_+_32978302 | 4.20 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr23_+_7379728 | 4.17 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr11_-_25418856 | 4.11 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr11_-_28614608 | 4.09 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr10_+_13209580 | 4.07 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr16_+_26449615 | 4.07 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr25_+_7982979 | 3.97 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr8_-_40464935 | 3.96 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr23_-_24682244 | 3.90 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr7_+_34290051 | 3.88 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr23_+_36115541 | 3.86 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr8_+_16004154 | 3.85 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr10_-_17484762 | 3.85 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr6_+_58915889 | 3.82 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr8_-_49431939 | 3.76 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr19_+_4912817 | 3.76 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr15_+_19838458 | 3.76 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr19_-_3381422 | 3.68 |
ENSDART00000105146
|
edn1
|
endothelin 1 |
| chr21_+_39941184 | 3.61 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr3_-_26204867 | 3.56 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr4_+_7508316 | 3.54 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr24_-_25691020 | 3.39 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr25_+_18564266 | 3.33 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr8_+_31872992 | 3.31 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr11_-_1400507 | 3.27 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr25_-_28674739 | 3.18 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr6_-_39275793 | 3.17 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr20_+_14941264 | 3.15 |
ENSDART00000080224
|
itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr8_-_18537866 | 3.15 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr2_+_4207209 | 3.04 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr17_-_42097267 | 3.03 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr20_-_25671342 | 2.90 |
ENSDART00000182775
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr9_+_17306162 | 2.86 |
ENSDART00000075926
|
scel
|
sciellin |
| chr18_+_48428713 | 2.84 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr20_+_40150612 | 2.84 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr3_+_40809892 | 2.83 |
ENSDART00000190803
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr20_+_33875256 | 2.81 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr18_+_27732285 | 2.81 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr5_+_34622320 | 2.80 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr2_+_33051730 | 2.75 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr8_-_40327397 | 2.72 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr13_+_22480496 | 2.71 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr1_+_36674584 | 2.70 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr13_+_22479988 | 2.70 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr8_+_37700090 | 2.64 |
ENSDART00000187885
ENSDART00000127633 |
adrb3a
|
adrenoceptor beta 3a |
| chr5_-_42272517 | 2.56 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_+_25708787 | 2.56 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr17_+_8175998 | 2.55 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr10_+_21650828 | 2.54 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr1_+_52563298 | 2.53 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr23_-_45501177 | 2.52 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr7_+_26138240 | 2.46 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr2_+_38261748 | 2.45 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr5_-_51166025 | 2.45 |
ENSDART00000097471
|
card9
|
caspase recruitment domain family, member 9 |
| chr3_+_32492467 | 2.44 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr3_+_24275766 | 2.43 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr14_+_35691889 | 2.43 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr14_-_32403554 | 2.43 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr16_+_13424534 | 2.42 |
ENSDART00000114944
|
zgc:101640
|
zgc:101640 |
| chr7_+_44445595 | 2.38 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr16_+_55059026 | 2.35 |
ENSDART00000109391
|
LO017815.1
|
Danio rerio nuclear receptor coactivator 7-like (LOC792958), mRNA. |
| chr16_-_36834505 | 2.35 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr14_+_30568961 | 2.35 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr23_-_33750135 | 2.34 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr21_-_25756119 | 2.29 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr7_+_46368520 | 2.19 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr23_+_27912079 | 2.18 |
ENSDART00000171859
|
BX539336.1
|
|
| chr7_+_48460239 | 2.17 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr22_-_11626014 | 2.16 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr21_+_19648814 | 2.00 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr23_-_5685023 | 1.99 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr13_-_37103126 | 1.97 |
ENSDART00000135935
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr14_-_28052474 | 1.94 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr8_-_14151051 | 1.90 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr6_-_609880 | 1.87 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr23_-_33750307 | 1.86 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr1_-_17569793 | 1.86 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr22_-_36856405 | 1.85 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr13_+_835390 | 1.85 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr1_-_17570013 | 1.77 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr18_-_16795262 | 1.76 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr18_-_42830563 | 1.76 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr16_+_54829574 | 1.74 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr8_-_39654669 | 1.72 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr16_+_54829293 | 1.71 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr21_+_39941875 | 1.68 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr17_-_26867725 | 1.65 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr11_+_31285127 | 1.64 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr14_+_26546175 | 1.63 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr1_+_41854298 | 1.61 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr25_-_29134000 | 1.60 |
ENSDART00000172027
ENSDART00000190447 |
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr2_-_24289641 | 1.59 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr25_-_13728111 | 1.59 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr23_+_26079467 | 1.58 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr11_-_6265574 | 1.55 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr1_-_38816685 | 1.53 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr6_+_8176486 | 1.53 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr23_-_6920107 | 1.52 |
ENSDART00000149230
ENSDART00000182946 |
si:ch211-117c9.2
|
si:ch211-117c9.2 |
| chr18_-_6803424 | 1.51 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr13_-_20381485 | 1.49 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr21_-_11820379 | 1.48 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr14_-_18672561 | 1.47 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr13_+_844150 | 1.45 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr18_+_48428126 | 1.44 |
ENSDART00000137978
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr24_-_2843107 | 1.43 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr3_+_16762483 | 1.42 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr1_+_26667872 | 1.40 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr10_-_43029001 | 1.38 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr13_+_23677949 | 1.36 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr2_+_33052169 | 1.35 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr24_-_21258945 | 1.34 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr7_+_69019851 | 1.33 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr7_-_41858513 | 1.29 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr20_-_26066020 | 1.29 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr20_+_33981946 | 1.29 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr2_-_51794472 | 1.28 |
ENSDART00000186652
|
BX908782.3
|
|
| chr1_-_8651718 | 1.28 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr21_+_39941559 | 1.27 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr21_+_12010505 | 1.26 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr16_-_24668620 | 1.24 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr4_+_16725960 | 1.22 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr17_-_26868169 | 1.18 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr6_-_35488180 | 1.16 |
ENSDART00000183258
|
rgs8
|
regulator of G protein signaling 8 |
| chr11_-_21528056 | 1.16 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr23_+_25856541 | 1.16 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr5_-_66301142 | 1.16 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr7_+_9904627 | 1.12 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr6_-_38418862 | 1.10 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr6_-_38419318 | 1.10 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr11_-_35575791 | 1.10 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr7_-_39203799 | 1.08 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr19_-_46777963 | 1.07 |
ENSDART00000046857
|
abrab
|
actin binding Rho activating protein b |
| chr8_+_26268100 | 1.04 |
ENSDART00000138882
|
slc26a6
|
solute carrier family 26, member 6 |
| chr9_+_11379782 | 1.02 |
ENSDART00000190282
ENSDART00000007308 |
wnt10a
|
wingless-type MMTV integration site family, member 10a |
| chr2_+_47927026 | 1.02 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr18_+_45228691 | 1.02 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr20_-_26420939 | 1.02 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr7_-_41851605 | 1.00 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr7_+_35075847 | 0.99 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr4_-_23858900 | 0.98 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr13_-_23031803 | 0.96 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr5_+_21211135 | 0.96 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr6_-_49078263 | 0.96 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr4_+_9030609 | 0.95 |
ENSDART00000154399
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr1_-_9109699 | 0.94 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr22_+_19111444 | 0.94 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr8_+_23485079 | 0.94 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr8_+_26268726 | 0.93 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr18_-_47533692 | 0.92 |
ENSDART00000191097
|
CABZ01112461.1
|
|
| chr20_-_26421112 | 0.90 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr24_+_29382109 | 0.89 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr4_-_20118468 | 0.89 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr13_+_45476181 | 0.87 |
ENSDART00000045329
|
mgst3b
|
microsomal glutathione S-transferase 3b |
| chr7_+_54642005 | 0.86 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr7_+_12835048 | 0.85 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr8_+_6576940 | 0.85 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr10_+_19525839 | 0.85 |
ENSDART00000162912
ENSDART00000158512 |
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr7_-_65236649 | 0.81 |
ENSDART00000185070
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr3_+_16449490 | 0.81 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr23_+_20408227 | 0.81 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr3_-_17716322 | 0.80 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr14_+_21686207 | 0.80 |
ENSDART00000034438
|
ran
|
RAN, member RAS oncogene family |
| chr1_+_29858032 | 0.80 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr14_+_36738069 | 0.80 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr14_+_17197132 | 0.79 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr12_+_6391243 | 0.78 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr20_+_46897504 | 0.76 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr6_-_40697585 | 0.76 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr9_-_45149695 | 0.75 |
ENSDART00000162033
|
CU467837.1
|
|
| chr19_+_8606883 | 0.73 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr16_-_17300030 | 0.72 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr16_+_25126935 | 0.69 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr17_+_39242437 | 0.68 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr15_-_568645 | 0.67 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr14_+_17382803 | 0.66 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr9_+_343062 | 0.66 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1b+gata2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 11.8 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 1.8 | 5.5 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 1.7 | 5.0 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.6 | 4.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 1.4 | 7.2 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 1.4 | 8.4 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 1.0 | 4.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 1.0 | 3.0 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 1.0 | 3.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.9 | 4.4 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.8 | 2.4 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.7 | 3.7 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.7 | 4.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.7 | 2.7 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.7 | 3.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.7 | 2.6 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 1.9 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.6 | 2.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.6 | 7.4 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.6 | 16.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.5 | 3.8 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.5 | 2.6 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 1.6 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.5 | 2.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.5 | 2.7 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 3.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 1.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.4 | 5.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.4 | 4.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.4 | 2.4 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
| 0.4 | 8.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 4.8 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.3 | 2.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 4.9 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.3 | 1.0 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 1.6 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.3 | 3.2 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.3 | 1.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 3.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.3 | 1.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 4.0 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 1.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 10.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.8 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.2 | 2.8 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.2 | 0.9 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 2.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 4.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 2.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 3.8 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 2.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 3.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 3.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 4.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 3.9 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 2.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 2.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 3.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 4.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 3.0 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.1 | 7.7 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 0.6 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 1.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 4.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 5.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 5.8 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.1 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.0 | 1.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 3.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0002031 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 2.2 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 1.0 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 2.9 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 11.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 3.6 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.9 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.4 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.9 | 2.8 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.7 | 5.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 8.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 4.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 4.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.3 | 1.3 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 9.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 3.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 5.9 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.2 | 2.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 6.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.2 | GO:0044327 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.1 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 6.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 8.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 4.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 48.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 3.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 4.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.8 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.7 | 3.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 8.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.7 | 2.6 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.6 | 3.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.6 | 7.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.5 | 2.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.5 | 4.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 2.5 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.5 | 5.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 3.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 6.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 14.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 5.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.4 | 2.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 2.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 5.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.3 | 3.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 3.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 1.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 1.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 7.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 2.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 0.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.3 | 2.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 5.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 2.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 5.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 4.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 4.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 2.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 2.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 3.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 3.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.9 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 1.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 9.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 1.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 4.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 4.2 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 4.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.6 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 1.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 10.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 2.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 2.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 3.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.0 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 8.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 6.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 4.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 13.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 5.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 2.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 2.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 3.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 8.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 3.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 2.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 13.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 8.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 6.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 3.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 3.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 16.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 4.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 7.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.6 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 3.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |