Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for foxm1
Z-value: 1.53
Transcription factors associated with foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxm1
|
ENSDARG00000003200 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxm1 | dr11_v1_chr4_-_5831036_5831036 | 0.82 | 6.9e-03 | Click! |
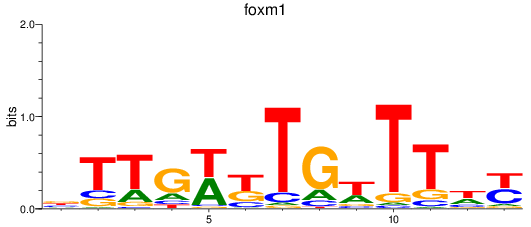
Activity profile of foxm1 motif
Sorted Z-values of foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4245902 | 2.57 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr9_+_33216945 | 2.36 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr13_+_31550185 | 1.62 |
ENSDART00000127843
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr16_+_25259313 | 1.57 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr22_+_835728 | 1.42 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr13_-_22961605 | 1.38 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr5_-_16475682 | 1.28 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr10_-_32465462 | 1.25 |
ENSDART00000134056
|
uvrag
|
UV radiation resistance associated gene |
| chr13_+_37653851 | 1.24 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr18_+_17534627 | 1.21 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr16_+_25068576 | 1.16 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr7_+_15736230 | 1.09 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr22_-_17671348 | 1.07 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr9_+_23770666 | 1.05 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr7_-_51775688 | 1.02 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr9_+_22780901 | 1.01 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr12_-_33558879 | 1.01 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr15_+_29393519 | 1.00 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr11_-_18017918 | 0.99 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr11_-_18017287 | 0.96 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr13_-_18011168 | 0.95 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr12_+_17154655 | 0.95 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr11_-_18107447 | 0.94 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr24_+_5893134 | 0.93 |
ENSDART00000077941
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr18_+_35128685 | 0.93 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr3_-_54607166 | 0.92 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr21_+_40685895 | 0.92 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr10_+_15454745 | 0.89 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr18_-_20458412 | 0.88 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr6_+_40922572 | 0.88 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_-_14040136 | 0.87 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr14_+_8638353 | 0.87 |
ENSDART00000163240
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr8_-_22558773 | 0.87 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr13_+_24679674 | 0.87 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr10_-_35257458 | 0.86 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr5_-_38197080 | 0.86 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr4_+_5196469 | 0.85 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr4_+_65127317 | 0.85 |
ENSDART00000166475
|
znf1126
|
zinc finger protein 1126 |
| chr15_-_1001177 | 0.84 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr2_-_44777592 | 0.83 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr11_+_44502410 | 0.82 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr10_-_35051691 | 0.81 |
ENSDART00000108670
ENSDART00000190711 |
supt20
|
SPT20 homolog, SAGA complex component |
| chr6_-_40922971 | 0.80 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr7_+_55950229 | 0.80 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr14_-_30905963 | 0.80 |
ENSDART00000183543
ENSDART00000186441 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr6_-_54444929 | 0.80 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr22_-_6066867 | 0.79 |
ENSDART00000142383
|
si:dkey-19a16.1
|
si:dkey-19a16.1 |
| chr19_-_38419575 | 0.79 |
ENSDART00000087639
|
smap2
|
small ArfGAP2 |
| chr2_+_43204919 | 0.78 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr17_+_32622933 | 0.78 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr25_+_28679672 | 0.78 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr3_-_60589292 | 0.78 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr3_+_1167026 | 0.77 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr4_-_5691257 | 0.77 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr19_+_25154066 | 0.77 |
ENSDART00000163220
|
si:ch211-239d6.2
|
si:ch211-239d6.2 |
| chr6_+_11397269 | 0.76 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr18_-_20458840 | 0.75 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr11_+_6881001 | 0.75 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr10_+_585719 | 0.75 |
ENSDART00000180167
|
smad4a
|
SMAD family member 4a |
| chr17_+_18810904 | 0.74 |
ENSDART00000130899
|
si:dkey-288a3.2
|
si:dkey-288a3.2 |
| chr1_-_51038885 | 0.73 |
ENSDART00000035150
|
spast
|
spastin |
| chr22_+_38935060 | 0.73 |
ENSDART00000183732
ENSDART00000130055 |
sirt7
|
sirtuin 7 |
| chr15_-_37589600 | 0.73 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr21_-_13856689 | 0.72 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr6_+_21536131 | 0.72 |
ENSDART00000113911
ENSDART00000188472 |
micall1a
|
MICAL-like 1a |
| chr4_-_39110934 | 0.71 |
ENSDART00000185041
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr19_-_7832439 | 0.70 |
ENSDART00000104703
ENSDART00000132336 |
znf687b
|
zinc finger protein 687b |
| chr15_-_23761580 | 0.68 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr5_+_29726428 | 0.68 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr3_+_32714157 | 0.68 |
ENSDART00000131774
|
setd1a
|
SET domain containing 1A |
| chr3_-_49138004 | 0.68 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr8_+_50531709 | 0.67 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr9_+_21259820 | 0.67 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr14_+_22498757 | 0.66 |
ENSDART00000021657
|
smyd5
|
SMYD family member 5 |
| chr1_+_52137528 | 0.65 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr17_-_25630822 | 0.65 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr7_-_13906409 | 0.64 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr5_-_18446483 | 0.64 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr15_+_1653779 | 0.64 |
ENSDART00000021299
|
nmd3
|
NMD3 ribosome export adaptor |
| chr21_+_3796620 | 0.64 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr7_+_17933384 | 0.64 |
ENSDART00000173701
|
mta2
|
metastasis associated 1 family, member 2 |
| chr3_-_29891218 | 0.64 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr1_-_23268013 | 0.64 |
ENSDART00000146575
|
rfc1
|
replication factor C (activator 1) 1 |
| chr10_-_42237304 | 0.63 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr13_+_11828516 | 0.63 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr10_-_244745 | 0.63 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr20_+_25904199 | 0.62 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr2_+_33926911 | 0.62 |
ENSDART00000109849
ENSDART00000135884 |
kif2c
|
kinesin family member 2C |
| chr2_-_24603325 | 0.62 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr4_-_39111612 | 0.62 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr1_+_45839927 | 0.59 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr14_-_41388178 | 0.59 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr9_+_27876146 | 0.59 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr16_+_40131473 | 0.58 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr11_+_30253968 | 0.58 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr11_+_30254556 | 0.58 |
ENSDART00000182316
|
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr7_-_19638319 | 0.57 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr2_+_11205795 | 0.57 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr10_+_22891126 | 0.57 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr12_-_23365737 | 0.57 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr17_-_21162821 | 0.56 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr25_+_3868438 | 0.56 |
ENSDART00000156438
|
tmem138
|
transmembrane protein 138 |
| chr2_-_29996036 | 0.56 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr6_+_28018390 | 0.55 |
ENSDART00000123324
ENSDART00000150915 |
sap130a
|
Sin3A-associated protein a |
| chr18_+_11858397 | 0.55 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr10_-_35052198 | 0.54 |
ENSDART00000147805
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr7_-_4125021 | 0.54 |
ENSDART00000167182
ENSDART00000173696 |
zgc:55733
|
zgc:55733 |
| chr15_-_25613114 | 0.54 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr14_-_52433292 | 0.54 |
ENSDART00000164272
|
rest
|
RE1-silencing transcription factor |
| chr7_-_6448182 | 0.54 |
ENSDART00000173045
|
si:ch1073-159d7.7
|
si:ch1073-159d7.7 |
| chr6_+_16949735 | 0.53 |
ENSDART00000155342
|
pimr13
|
Pim proto-oncogene, serine/threonine kinase, related 13 |
| chr3_+_53116172 | 0.53 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr25_-_24202576 | 0.53 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr5_+_39099380 | 0.53 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr18_-_42333428 | 0.53 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr16_-_31445781 | 0.52 |
ENSDART00000056551
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr1_+_19538299 | 0.51 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr15_-_28587147 | 0.51 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr3_+_60589157 | 0.51 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr5_+_27267186 | 0.51 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr13_+_33298338 | 0.51 |
ENSDART00000131892
ENSDART00000143895 |
iqcc
|
IQ motif containing C |
| chr19_+_7759354 | 0.50 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr13_+_7578111 | 0.50 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr19_-_6134802 | 0.50 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_-_6444011 | 0.50 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr17_+_8799661 | 0.50 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr23_+_39611688 | 0.49 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr11_-_42418374 | 0.48 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr10_+_573667 | 0.48 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr1_-_55166511 | 0.48 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr12_+_28888975 | 0.47 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr10_-_38243579 | 0.47 |
ENSDART00000150159
|
usp25
|
ubiquitin specific peptidase 25 |
| chr2_+_31838442 | 0.47 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr18_+_26829086 | 0.46 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr10_+_35257651 | 0.46 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr25_-_35045250 | 0.46 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr9_-_55772937 | 0.46 |
ENSDART00000159192
|
akap17a
|
A kinase (PRKA) anchor protein 17A |
| chr22_-_22301672 | 0.45 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr19_-_10324182 | 0.45 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr7_+_52766211 | 0.45 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr18_+_44768829 | 0.45 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr8_+_40210398 | 0.44 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr13_+_2894536 | 0.44 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr11_-_11336986 | 0.44 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr19_-_11966015 | 0.43 |
ENSDART00000123409
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr7_+_17255705 | 0.43 |
ENSDART00000053357
ENSDART00000167849 ENSDART00000165833 |
nitr9
si:ch73-46n24.1
|
novel immune-type receptor 9 si:ch73-46n24.1 |
| chr3_+_37112693 | 0.43 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr15_-_23692359 | 0.42 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr14_-_22015232 | 0.42 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr14_+_52481288 | 0.42 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr3_+_1107102 | 0.42 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr11_+_45153104 | 0.41 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr20_-_22798794 | 0.41 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr7_+_30282389 | 0.40 |
ENSDART00000108782
|
polr2m
|
RNA polymerase II subunit M |
| chr7_-_19642417 | 0.40 |
ENSDART00000160936
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr13_+_10023256 | 0.40 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr15_+_23534126 | 0.39 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr19_+_4139065 | 0.38 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr4_+_9011825 | 0.38 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr11_-_2250767 | 0.38 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr12_-_9516981 | 0.38 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr3_-_50118140 | 0.38 |
ENSDART00000131913
|
hgs
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr8_+_23147218 | 0.38 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr6_+_40523370 | 0.38 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr16_+_1100559 | 0.37 |
ENSDART00000092657
|
adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr20_+_9124369 | 0.37 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr1_-_18585046 | 0.37 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr9_+_2522797 | 0.36 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr3_-_13461361 | 0.36 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr21_-_23046606 | 0.36 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr18_+_41560822 | 0.36 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr16_+_46410520 | 0.35 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr3_-_13461056 | 0.35 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr5_-_23596339 | 0.34 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr7_+_38811800 | 0.34 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr17_-_11439815 | 0.33 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr14_-_4177311 | 0.33 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr17_+_23975762 | 0.33 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr25_-_20730800 | 0.33 |
ENSDART00000172170
|
si:ch211-127m7.2
|
si:ch211-127m7.2 |
| chr8_-_18211605 | 0.33 |
ENSDART00000114177
|
CT025742.1
|
|
| chr17_-_8656155 | 0.33 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr8_-_21071476 | 0.32 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr13_+_12761707 | 0.32 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr11_-_24347644 | 0.31 |
ENSDART00000089777
|
si:ch211-15p9.2
|
si:ch211-15p9.2 |
| chr25_+_36045072 | 0.31 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr7_-_50604626 | 0.31 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr5_-_13251907 | 0.31 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr15_-_28082310 | 0.31 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr5_-_51902935 | 0.31 |
ENSDART00000182160
|
mtx3
|
metaxin 3 |
| chr14_+_31865099 | 0.31 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr22_+_2254972 | 0.31 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr12_-_19119176 | 0.30 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr18_-_37407235 | 0.30 |
ENSDART00000132315
|
cep126
|
centrosomal protein 126 |
| chr4_-_23759192 | 0.30 |
ENSDART00000014685
ENSDART00000131690 |
cpm
|
carboxypeptidase M |
| chr14_+_44804326 | 0.29 |
ENSDART00000079866
ENSDART00000172974 |
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr3_-_19561058 | 0.29 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr17_+_10593398 | 0.29 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr25_+_4812685 | 0.29 |
ENSDART00000193103
|
myo5c
|
myosin VC |
| chr22_+_2315996 | 0.29 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr5_-_67757188 | 0.29 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.5 | 2.6 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 1.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 0.9 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.3 | 1.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 0.7 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 1.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 1.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.8 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 0.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 1.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 0.6 | GO:1904589 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.6 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.7 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.4 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.6 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.4 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 1.0 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 0.3 | GO:0071043 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.9 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 1.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.8 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.6 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.7 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 1.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.4 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.0 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.7 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.2 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 0.9 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) limb morphogenesis(GO:0035108) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.2 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.4 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.4 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 1.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.8 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.0 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 1.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.8 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.4 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 3.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 0.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.6 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.2 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.4 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.1 | 1.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 1.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |