Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for foxk1_foxj3
Z-value: 0.86
Transcription factors associated with foxk1_foxj3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
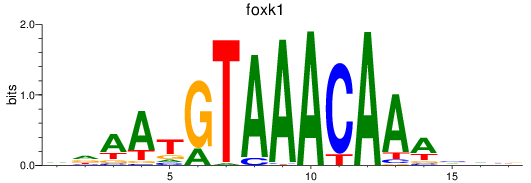
foxk1
|
ENSDARG00000037872 | forkhead box K1 |
|
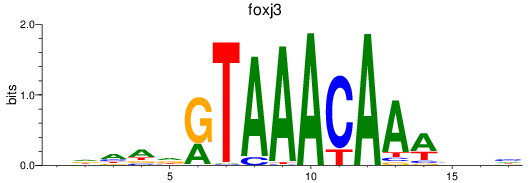
foxj3
|
ENSDARG00000075774 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxk1 | dr11_v1_chr3_-_40933415_40933415 | 0.90 | 1.0e-03 | Click! |
| foxj3 | dr11_v1_chr11_-_26362294_26362334 | 0.75 | 1.9e-02 | Click! |
Activity profile of foxk1_foxj3 motif
Sorted Z-values of foxk1_foxj3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_15155684 | 2.21 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr15_+_19990068 | 2.00 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr7_+_46019780 | 1.84 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr17_+_44030692 | 1.44 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr13_-_33207367 | 1.41 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr16_-_47381519 | 1.36 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr10_-_14929392 | 1.27 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr23_-_1571682 | 1.17 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr17_+_14965570 | 1.11 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr11_+_11120532 | 1.11 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr22_+_38276024 | 1.10 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr20_-_52928541 | 1.10 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr16_-_26820634 | 1.08 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_-_2690083 | 1.07 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr25_-_13871118 | 1.06 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr5_+_44944778 | 1.06 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr13_+_29925397 | 1.05 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr10_-_14929630 | 1.03 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr4_-_20135406 | 1.01 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr12_-_28537615 | 1.00 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr5_+_22836364 | 0.98 |
ENSDART00000131885
|
si:ch211-26b3.2
|
si:ch211-26b3.2 |
| chr11_+_2710530 | 0.96 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr1_+_53321878 | 0.96 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr5_-_66749535 | 0.92 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr7_-_46019756 | 0.91 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr2_+_16696052 | 0.90 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr16_-_41714988 | 0.89 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr3_-_25369557 | 0.88 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr20_+_46741074 | 0.85 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr5_+_55984270 | 0.85 |
ENSDART00000047358
ENSDART00000138191 |
fkbp6
|
FK506 binding protein 6 |
| chr21_-_13661631 | 0.85 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr24_-_19718077 | 0.85 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr11_-_25257045 | 0.85 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr9_+_41459759 | 0.83 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr21_-_13662237 | 0.83 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr22_-_28777557 | 0.83 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr20_-_9428021 | 0.82 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr17_+_24111392 | 0.81 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr15_+_34069746 | 0.80 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr6_-_8466717 | 0.80 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr5_-_38384289 | 0.79 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr1_-_9277986 | 0.78 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr25_-_13408760 | 0.78 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr4_-_20135919 | 0.77 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr19_-_3821678 | 0.76 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr16_-_49646625 | 0.74 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr21_-_43398122 | 0.74 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr11_-_25257595 | 0.73 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr5_-_4931266 | 0.73 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr16_+_25011994 | 0.71 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr3_+_53116172 | 0.71 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr14_-_33095917 | 0.70 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr9_+_24088062 | 0.69 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr8_+_16990120 | 0.68 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_40922971 | 0.68 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr5_-_68058168 | 0.67 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr12_-_5998898 | 0.67 |
ENSDART00000142659
ENSDART00000004896 |
kat7b
|
K(lysine) acetyltransferase 7b |
| chr2_-_24603325 | 0.67 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr1_+_12394205 | 0.66 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr6_-_9565526 | 0.65 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr4_-_1720648 | 0.65 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr9_-_12885201 | 0.63 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr2_-_4787566 | 0.63 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr16_+_9580699 | 0.62 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr20_+_53368611 | 0.62 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr23_+_7692042 | 0.61 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr14_+_35424539 | 0.61 |
ENSDART00000171809
ENSDART00000162185 |
sytl4
|
synaptotagmin-like 4 |
| chr8_+_37749263 | 0.60 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr21_-_7940043 | 0.60 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr2_+_27386617 | 0.59 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr16_-_12097558 | 0.58 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr11_+_11030908 | 0.58 |
ENSDART00000111091
|
pla2r1
|
phospholipase A2 receptor 1 |
| chr14_-_33177935 | 0.55 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr16_-_12097394 | 0.55 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr10_+_573667 | 0.55 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr3_-_26191960 | 0.55 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr5_+_36611128 | 0.55 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr11_+_25430851 | 0.54 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr7_-_37555208 | 0.54 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr22_-_34979139 | 0.53 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr15_+_25681044 | 0.53 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr16_+_26732086 | 0.52 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr8_-_39859688 | 0.52 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr16_+_28578352 | 0.52 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr5_-_12743196 | 0.51 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr5_-_1827575 | 0.51 |
ENSDART00000171193
|
mcc
|
mutated in colorectal cancers |
| chr2_+_27330461 | 0.51 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr5_+_8196264 | 0.50 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr22_-_29689981 | 0.50 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr12_-_32066469 | 0.50 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr3_+_19685873 | 0.49 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr24_-_6678640 | 0.49 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr22_-_28777374 | 0.48 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr2_-_17492486 | 0.48 |
ENSDART00000189464
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr21_+_20386865 | 0.48 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr8_+_4803906 | 0.48 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr16_+_48714048 | 0.48 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr3_+_42999277 | 0.47 |
ENSDART00000163595
|
ints1
|
integrator complex subunit 1 |
| chr5_-_40190949 | 0.47 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr18_-_5875433 | 0.47 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr15_-_28805493 | 0.47 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr19_+_25971000 | 0.47 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr11_-_13341051 | 0.47 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr5_+_50371509 | 0.46 |
ENSDART00000167163
|
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr2_-_42109575 | 0.46 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr8_+_26034623 | 0.46 |
ENSDART00000004521
ENSDART00000142555 |
arih2
|
ariadne homolog 2 (Drosophila) |
| chr17_+_24036791 | 0.46 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr7_+_24528866 | 0.45 |
ENSDART00000180552
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr9_+_21306902 | 0.45 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr13_-_48431766 | 0.45 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr6_-_39700965 | 0.45 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr8_-_29822527 | 0.45 |
ENSDART00000167487
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr23_-_18707418 | 0.45 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr16_+_38350704 | 0.45 |
ENSDART00000140761
|
elp6
|
elongator acetyltransferase complex subunit 6 |
| chr8_-_1267247 | 0.44 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr3_+_42999844 | 0.44 |
ENSDART00000183950
|
ints1
|
integrator complex subunit 1 |
| chr17_+_4325693 | 0.44 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr5_+_44846434 | 0.44 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr13_+_29926094 | 0.44 |
ENSDART00000057528
|
cuedc2
|
CUE domain containing 2 |
| chr15_-_37589600 | 0.44 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr1_-_12394048 | 0.43 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr18_+_15271993 | 0.43 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr20_+_25904199 | 0.43 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr3_+_55031685 | 0.43 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr13_+_12761707 | 0.42 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr6_+_4255319 | 0.41 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr16_+_40954481 | 0.41 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr11_+_18612421 | 0.41 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr12_-_22509069 | 0.40 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr21_+_17051478 | 0.40 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr11_-_13341483 | 0.40 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr12_+_30726425 | 0.40 |
ENSDART00000153275
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr2_+_1988036 | 0.39 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr1_-_23308225 | 0.39 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr3_-_60711127 | 0.38 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr22_-_29689485 | 0.38 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr20_+_42565049 | 0.38 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr18_-_13121983 | 0.37 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr7_+_40205394 | 0.37 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr7_-_13906409 | 0.37 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr5_-_51998708 | 0.36 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr16_-_30847192 | 0.36 |
ENSDART00000191106
ENSDART00000128158 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr13_-_25842074 | 0.36 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr1_+_51191049 | 0.36 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr15_+_39977461 | 0.36 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr11_+_6295370 | 0.35 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr15_+_26933196 | 0.35 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr5_-_24008997 | 0.35 |
ENSDART00000066645
|
eif1axa
|
eukaryotic translation initiation factor 1A, X-linked, a |
| chr13_-_5257303 | 0.35 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr19_-_22843480 | 0.35 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr24_+_7322116 | 0.35 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr14_-_34633960 | 0.34 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr12_+_8822717 | 0.34 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr22_+_26798853 | 0.34 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr23_+_31596194 | 0.34 |
ENSDART00000160748
|
tbpl1
|
TBP-like 1 |
| chr23_+_31596441 | 0.34 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr10_-_38243579 | 0.33 |
ENSDART00000150159
|
usp25
|
ubiquitin specific peptidase 25 |
| chr9_+_2343096 | 0.33 |
ENSDART00000062292
ENSDART00000191722 ENSDART00000135180 |
atf2
|
activating transcription factor 2 |
| chr25_+_2263857 | 0.33 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr2_-_26720854 | 0.33 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr7_+_19851422 | 0.33 |
ENSDART00000142970
ENSDART00000190027 |
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr17_-_7218481 | 0.32 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr1_-_23370395 | 0.32 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr15_-_7337148 | 0.31 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr18_+_5875268 | 0.31 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr13_-_23665580 | 0.31 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr9_-_30494727 | 0.31 |
ENSDART00000148729
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr10_-_36633882 | 0.31 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr23_+_32029304 | 0.31 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr23_-_45398622 | 0.31 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr18_+_20047374 | 0.31 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr24_+_19542323 | 0.31 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr10_+_2529037 | 0.30 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr12_+_45238292 | 0.30 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr18_-_6855991 | 0.30 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr17_+_37215820 | 0.30 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr21_-_41588129 | 0.29 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr21_-_27273147 | 0.29 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr23_+_21978584 | 0.29 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr15_-_30984557 | 0.29 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr25_-_25142387 | 0.29 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr7_+_5530154 | 0.29 |
ENSDART00000181610
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr2_-_23677422 | 0.29 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr21_-_27272657 | 0.28 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr18_+_21113216 | 0.28 |
ENSDART00000060191
|
kmt5b
|
lysine methyltransferase 5B |
| chr9_-_27868267 | 0.28 |
ENSDART00000079502
|
dbr1
|
debranching RNA lariats 1 |
| chr17_-_14966384 | 0.28 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr19_-_47997424 | 0.28 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr5_-_50371205 | 0.27 |
ENSDART00000157800
|
si:ch73-280o22.2
|
si:ch73-280o22.2 |
| chr10_+_31809226 | 0.27 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr23_+_32028574 | 0.27 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr11_-_8126223 | 0.27 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr1_+_35494837 | 0.26 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr5_-_32074534 | 0.26 |
ENSDART00000112342
ENSDART00000135855 |
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr8_+_10339869 | 0.26 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr5_-_25236340 | 0.26 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr17_-_23709347 | 0.26 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr5_-_23485161 | 0.26 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr24_-_5932982 | 0.26 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr20_+_52774730 | 0.26 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxk1_foxj3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.4 | 1.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.3 | 1.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 1.1 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 1.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.8 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 1.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.3 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.3 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.2 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 2.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.5 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 1.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.8 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of hormone metabolic process(GO:0032350) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.2 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 1.2 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.3 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.4 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 2.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 1.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 1.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.4 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 2.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |