Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
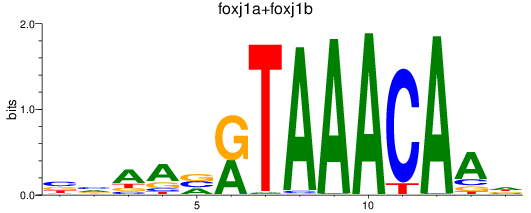
Results for foxj1a+foxj1b
Z-value: 0.77
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1a | dr11_v1_chr3_+_60716904_60716904 | -0.13 | 7.3e-01 | Click! |
| foxj1b | dr11_v1_chr12_+_20149707_20149707 | -0.08 | 8.5e-01 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_54014539 | 0.84 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr10_-_35257458 | 0.47 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr6_-_14038804 | 0.46 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr9_+_41459759 | 0.45 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr20_-_54014373 | 0.45 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr24_+_39211288 | 0.43 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr17_-_4252221 | 0.43 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr15_+_19990068 | 0.43 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr14_+_26439227 | 0.42 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr23_+_38159715 | 0.42 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr16_+_26732086 | 0.41 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr1_+_47178529 | 0.39 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr4_-_20135406 | 0.38 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr10_-_3416258 | 0.38 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr10_+_35257651 | 0.37 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr10_-_14929392 | 0.36 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr13_-_7233811 | 0.36 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr20_+_53368611 | 0.36 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr12_+_3912544 | 0.36 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr23_-_1571682 | 0.36 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr2_+_38271392 | 0.35 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr19_-_10915898 | 0.35 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr4_-_12795436 | 0.35 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr19_+_4968947 | 0.35 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr13_+_29925397 | 0.34 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr8_+_23034718 | 0.34 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr1_+_51537250 | 0.33 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr5_-_38342992 | 0.33 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr14_+_15155684 | 0.33 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr11_+_11120532 | 0.32 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr5_+_44944778 | 0.32 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr6_-_9695294 | 0.32 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr22_+_3184500 | 0.32 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr2_+_16696052 | 0.31 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr1_+_53321878 | 0.31 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr8_-_39859688 | 0.31 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr15_-_28805493 | 0.30 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr16_-_49646625 | 0.30 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr16_-_31435020 | 0.30 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr13_+_29926094 | 0.30 |
ENSDART00000057528
|
cuedc2
|
CUE domain containing 2 |
| chr6_+_32834760 | 0.28 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr4_-_20135919 | 0.28 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr16_+_9580699 | 0.28 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr22_+_7462997 | 0.28 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr9_+_2041535 | 0.28 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr25_-_10571078 | 0.27 |
ENSDART00000153898
|
si:ch211-107e6.5
|
si:ch211-107e6.5 |
| chr6_-_32834385 | 0.27 |
ENSDART00000129803
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr14_-_28566238 | 0.27 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr18_-_5875433 | 0.27 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr21_+_3796620 | 0.27 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr6_-_40922971 | 0.27 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr21_-_30030644 | 0.27 |
ENSDART00000190810
|
CU855895.2
|
|
| chr15_+_34069746 | 0.27 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr13_-_33207367 | 0.26 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr10_-_42108137 | 0.26 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr22_-_28777557 | 0.26 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr8_+_49065348 | 0.26 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr3_-_49504023 | 0.26 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr5_+_29726428 | 0.25 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr15_-_7337148 | 0.25 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr6_+_40922572 | 0.25 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_-_20866758 | 0.25 |
ENSDART00000165374
|
tpra
|
translocated promoter region a, nuclear basket protein |
| chr3_-_40664868 | 0.25 |
ENSDART00000138783
ENSDART00000178567 |
rnf216
|
ring finger protein 216 |
| chr23_+_7692042 | 0.25 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr1_+_9153141 | 0.25 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr15_-_7337537 | 0.25 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr2_+_20866898 | 0.25 |
ENSDART00000150086
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr24_+_19542323 | 0.25 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr25_-_34740627 | 0.24 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr10_-_35186310 | 0.24 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr13_+_18533005 | 0.24 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr18_+_20226843 | 0.24 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr15_+_26933196 | 0.24 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr7_-_38570878 | 0.24 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr21_+_17051478 | 0.23 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr13_-_31687925 | 0.23 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr19_+_4856351 | 0.23 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr7_-_51300277 | 0.23 |
ENSDART00000174174
|
gc2
|
guanylyl cyclase 2 |
| chr10_-_14929630 | 0.23 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr6_-_41138854 | 0.23 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr4_+_5868034 | 0.23 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr7_+_36467315 | 0.22 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr2_-_10631767 | 0.22 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr21_-_30031396 | 0.21 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr17_-_20167206 | 0.21 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr9_-_12885201 | 0.21 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr17_+_14965570 | 0.21 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr21_-_3853204 | 0.21 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr22_+_10232527 | 0.21 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr16_-_26820634 | 0.21 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr23_-_45398622 | 0.21 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr5_+_37504309 | 0.21 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr3_-_36419641 | 0.21 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr1_-_23268013 | 0.20 |
ENSDART00000146575
|
rfc1
|
replication factor C (activator 1) 1 |
| chr4_+_11723852 | 0.20 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr20_-_2667902 | 0.20 |
ENSDART00000036373
|
cfap206
|
cilia and flagella associated protein 206 |
| chr2_+_41524238 | 0.20 |
ENSDART00000122860
ENSDART00000017977 |
acvr1l
|
activin A receptor, type 1 like |
| chr23_+_9508538 | 0.20 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr17_-_2690083 | 0.20 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr15_+_39977461 | 0.20 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr3_+_33440615 | 0.20 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr19_+_25971000 | 0.19 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr10_-_23099809 | 0.19 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr2_-_26720854 | 0.19 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr4_-_12795030 | 0.19 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr13_+_28690355 | 0.19 |
ENSDART00000137475
ENSDART00000128246 |
polr1c
|
polymerase (RNA) I polypeptide C |
| chr2_-_42958113 | 0.19 |
ENSDART00000139945
|
oc90
|
otoconin 90 |
| chr2_+_1988036 | 0.19 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr5_-_66160415 | 0.19 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr2_+_19195841 | 0.19 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr21_-_11632403 | 0.19 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr5_+_36895545 | 0.19 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr20_-_18794789 | 0.18 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr24_+_14240196 | 0.18 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr23_+_33296588 | 0.18 |
ENSDART00000030368
|
si:ch211-226m16.3
|
si:ch211-226m16.3 |
| chr2_-_17492080 | 0.18 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr13_-_25842074 | 0.18 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr21_+_3796196 | 0.18 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr3_-_57630791 | 0.18 |
ENSDART00000129598
|
usp36
|
ubiquitin specific peptidase 36 |
| chr16_-_41714988 | 0.18 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr20_-_2641233 | 0.18 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr2_-_23677422 | 0.18 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr19_+_619200 | 0.18 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr3_-_31057624 | 0.18 |
ENSDART00000152901
|
armc5
|
armadillo repeat containing 5 |
| chr5_-_24231139 | 0.17 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr14_-_33095917 | 0.17 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr7_+_36467796 | 0.17 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr3_-_25369557 | 0.17 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr1_-_9277986 | 0.17 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr14_+_30272891 | 0.17 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr8_-_35960987 | 0.17 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr14_-_41478265 | 0.17 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr17_+_24111392 | 0.16 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr12_+_18899396 | 0.16 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr16_+_33121260 | 0.16 |
ENSDART00000058472
|
akirin1
|
akirin 1 |
| chr22_+_39007533 | 0.16 |
ENSDART00000185958
ENSDART00000129848 |
fam208aa
|
family with sequence similarity 208, member Aa |
| chr18_-_45761868 | 0.16 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr10_-_42237304 | 0.16 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr24_-_25461267 | 0.16 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr1_+_35494837 | 0.16 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr12_-_22509069 | 0.16 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr5_-_19014589 | 0.15 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr25_+_19734038 | 0.15 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr22_-_28777374 | 0.15 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr22_+_18166660 | 0.15 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr17_+_10094063 | 0.15 |
ENSDART00000168055
|
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr20_+_42565049 | 0.15 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr17_-_28100501 | 0.15 |
ENSDART00000149543
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr11_+_18612421 | 0.15 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr16_-_21692024 | 0.15 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr5_-_66749535 | 0.15 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr2_+_27330461 | 0.15 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr18_-_44935174 | 0.14 |
ENSDART00000081025
|
pex16
|
peroxisomal biogenesis factor 16 |
| chr25_-_13871118 | 0.14 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr5_+_36895860 | 0.14 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr11_-_16394971 | 0.14 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr16_+_33121106 | 0.14 |
ENSDART00000110195
|
akirin1
|
akirin 1 |
| chr23_+_31596441 | 0.14 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr7_+_38808027 | 0.14 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr19_+_20177887 | 0.14 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr8_-_1267247 | 0.14 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr2_-_31791633 | 0.14 |
ENSDART00000180662
|
retreg1
|
reticulophagy regulator 1 |
| chr25_+_3868438 | 0.14 |
ENSDART00000156438
|
tmem138
|
transmembrane protein 138 |
| chr3_+_55031685 | 0.14 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr14_-_21661015 | 0.14 |
ENSDART00000189403
ENSDART00000172442 ENSDART00000181913 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr22_-_22231720 | 0.14 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr19_+_40250682 | 0.14 |
ENSDART00000102888
|
cdk6
|
cyclin-dependent kinase 6 |
| chr6_-_37745508 | 0.13 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr17_-_25630822 | 0.13 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr25_+_28555584 | 0.13 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr7_-_28611145 | 0.13 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr13_+_47821524 | 0.13 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr23_+_24598910 | 0.13 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr11_-_45420212 | 0.13 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr6_-_10728057 | 0.13 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr7_-_50367326 | 0.13 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr5_+_40299568 | 0.12 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr24_-_19718077 | 0.12 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr18_-_24988645 | 0.12 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_+_23099890 | 0.12 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr14_-_33425170 | 0.12 |
ENSDART00000124629
ENSDART00000105800 ENSDART00000001318 |
nkap
|
NFKB activating protein |
| chr12_-_28537615 | 0.12 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr3_+_32671146 | 0.12 |
ENSDART00000039466
|
rnf25
|
ring finger protein 25 |
| chr2_-_17492486 | 0.12 |
ENSDART00000189464
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr10_+_22918338 | 0.12 |
ENSDART00000167874
ENSDART00000171298 |
zgc:103508
|
zgc:103508 |
| chr18_-_17399291 | 0.12 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr20_+_16173618 | 0.12 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr11_+_16152316 | 0.12 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr20_-_48061351 | 0.12 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr7_+_13830052 | 0.12 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr24_+_9298198 | 0.12 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr19_-_22843480 | 0.12 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr23_-_42876596 | 0.12 |
ENSDART00000086156
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr20_+_46741074 | 0.12 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr16_+_25608778 | 0.11 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr23_-_27608257 | 0.11 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr24_+_17069420 | 0.11 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr8_+_4803906 | 0.11 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr7_-_37555208 | 0.11 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr6_-_18228358 | 0.11 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 0.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.3 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.4 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.5 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.5 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |