Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for foxi1+foxi2+foxi3a+foxi3b
Z-value: 0.78
Transcription factors associated with foxi1+foxi2+foxi3a+foxi3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxi3b
|
ENSDARG00000009550 | forkhead box I3b |
|
foxi3a
|
ENSDARG00000055926 | forkhead box I3a |
|
foxi2
|
ENSDARG00000069715 | forkhead box I2 |
|
foxi1
|
ENSDARG00000104566 | forkhead box i1 |
|
foxi3b
|
ENSDARG00000112022 | forkhead box I3b |
|
foxi3b
|
ENSDARG00000112303 | forkhead box I3b |
|
foxi2
|
ENSDARG00000116791 | forkhead box I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxi3a | dr11_v1_chr21_+_30351256_30351256 | -0.85 | 3.5e-03 | Click! |
| foxi3b | dr11_v1_chr14_+_34514336_34514336 | -0.73 | 2.6e-02 | Click! |
| foxi2 | dr11_v1_chr13_+_13578552_13578552 | -0.71 | 3.1e-02 | Click! |
| foxi1 | dr11_v1_chr12_-_43664682_43664682 | -0.30 | 4.3e-01 | Click! |
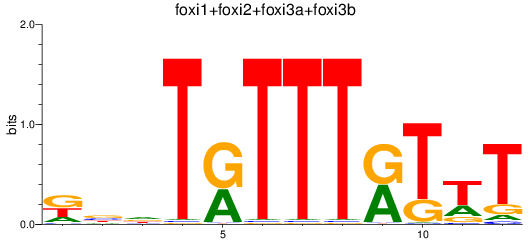
Activity profile of foxi1+foxi2+foxi3a+foxi3b motif
Sorted Z-values of foxi1+foxi2+foxi3a+foxi3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_33647138 | 1.85 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr15_+_38299385 | 1.46 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr15_+_38299563 | 1.38 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr22_+_23359369 | 1.32 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr5_+_36768674 | 1.24 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr4_-_16628801 | 1.20 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr13_+_37653851 | 1.19 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr10_-_14929392 | 1.04 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr19_+_29303847 | 0.95 |
ENSDART00000009149
|
maco1a
|
macoilin 1a |
| chr7_+_13830052 | 0.93 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr22_-_28777557 | 0.92 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr3_-_2623176 | 0.87 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr8_+_8643901 | 0.78 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr6_+_4255319 | 0.75 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr5_-_49951106 | 0.75 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr1_-_6028876 | 0.72 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr3_-_60589292 | 0.71 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr18_+_27077853 | 0.69 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr23_-_42810664 | 0.68 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr4_+_11723852 | 0.66 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr21_+_4116437 | 0.66 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr19_+_1510971 | 0.65 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr8_+_10862353 | 0.64 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr7_+_52766211 | 0.63 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr8_+_13368150 | 0.62 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr17_-_2685026 | 0.62 |
ENSDART00000191014
ENSDART00000179309 |
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr20_+_42565049 | 0.61 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr19_-_2822372 | 0.59 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr17_+_14965570 | 0.59 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr7_-_33868903 | 0.59 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr4_-_20135406 | 0.58 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr14_+_32926385 | 0.58 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr17_-_11417904 | 0.57 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_-_49800755 | 0.56 |
ENSDART00000180072
|
fjx1
|
four jointed box 1 |
| chr16_+_38820486 | 0.56 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr15_+_23657051 | 0.55 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr2_-_31798717 | 0.55 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr2_-_51507540 | 0.54 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr7_-_49801183 | 0.54 |
ENSDART00000052083
|
fjx1
|
four jointed box 1 |
| chr22_-_28777374 | 0.53 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr4_-_5691257 | 0.53 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr3_-_55537096 | 0.53 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr11_+_42641404 | 0.52 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr17_-_11417551 | 0.52 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_+_31437547 | 0.51 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr10_+_1849874 | 0.50 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr17_-_29271359 | 0.50 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr14_+_15231097 | 0.50 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr9_+_27720428 | 0.50 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr17_+_44756247 | 0.50 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr16_+_16265850 | 0.49 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr20_-_31252809 | 0.49 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr15_-_14038227 | 0.47 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr10_-_7555326 | 0.47 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr24_-_18659147 | 0.47 |
ENSDART00000166039
|
zgc:66014
|
zgc:66014 |
| chr7_+_15736230 | 0.46 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr4_-_20135919 | 0.46 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr13_+_50778187 | 0.45 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr17_+_25856671 | 0.45 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr1_+_51191049 | 0.45 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_-_33215261 | 0.45 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr24_-_9002038 | 0.45 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr9_-_27719998 | 0.44 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr1_+_45839927 | 0.44 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr2_-_57110477 | 0.44 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr11_+_16574625 | 0.44 |
ENSDART00000104072
|
kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr8_-_2616326 | 0.43 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr1_+_9153141 | 0.43 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr18_-_21725638 | 0.43 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr10_-_11840353 | 0.42 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr19_-_29303788 | 0.42 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr22_+_10543329 | 0.42 |
ENSDART00000091850
|
atrip
|
ATR interacting protein |
| chr3_+_53116172 | 0.42 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr18_+_37015185 | 0.41 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr14_-_16772452 | 0.41 |
ENSDART00000137493
|
mrnip
|
MRN complex interacting protein |
| chr10_-_7555660 | 0.41 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr15_+_25681044 | 0.41 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr9_-_41040492 | 0.41 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr3_-_33113879 | 0.41 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr5_-_2676783 | 0.40 |
ENSDART00000159661
|
CABZ01072548.1
|
|
| chr20_-_45062514 | 0.40 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr8_-_18316482 | 0.40 |
ENSDART00000190156
|
rnf220b
|
ring finger protein 220b |
| chr5_+_61361815 | 0.40 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr5_+_24063046 | 0.39 |
ENSDART00000051548
ENSDART00000133355 ENSDART00000142268 |
gps2
|
G protein pathway suppressor 2 |
| chr19_+_25971000 | 0.39 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr25_+_6451038 | 0.39 |
ENSDART00000009971
|
snx33
|
sorting nexin 33 |
| chr16_-_41488023 | 0.39 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr13_+_24679674 | 0.38 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr2_-_10631767 | 0.38 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr3_+_60589157 | 0.38 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr9_+_2762270 | 0.38 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr5_+_36666715 | 0.38 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr11_+_25693395 | 0.37 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr9_+_24088062 | 0.37 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr9_-_12574473 | 0.37 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr4_+_6833735 | 0.37 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr23_+_22873415 | 0.37 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr25_-_14087377 | 0.36 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr17_+_37511092 | 0.36 |
ENSDART00000187935
|
mta1
|
metastasis associated 1 |
| chr12_+_34854562 | 0.36 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr8_-_7567815 | 0.35 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr7_+_52712807 | 0.35 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr16_-_41487589 | 0.35 |
ENSDART00000188115
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr5_-_33255759 | 0.34 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr5_-_48070779 | 0.34 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr2_+_26060528 | 0.34 |
ENSDART00000058111
|
grin3ba
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Ba |
| chr22_+_30112390 | 0.34 |
ENSDART00000191715
|
add3a
|
adducin 3 (gamma) a |
| chr19_-_45650994 | 0.34 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr15_+_26940569 | 0.34 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr7_-_37555208 | 0.33 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr17_+_32622933 | 0.33 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr16_+_5612547 | 0.33 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr17_+_50261603 | 0.33 |
ENSDART00000154503
ENSDART00000154467 |
syncripl
|
synaptotagmin binding, cytoplasmic RNA interacting protein, like |
| chr19_-_9503473 | 0.32 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr16_-_54971277 | 0.32 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr17_+_37510834 | 0.32 |
ENSDART00000045583
|
mta1
|
metastasis associated 1 |
| chr5_-_4931266 | 0.32 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr16_+_25608778 | 0.32 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr23_-_33680265 | 0.31 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr3_+_22432069 | 0.31 |
ENSDART00000123054
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr22_-_10752471 | 0.31 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr6_+_52918537 | 0.31 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr6_-_40922971 | 0.30 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr6_-_12270226 | 0.30 |
ENSDART00000180473
|
pkp4
|
plakophilin 4 |
| chr5_+_29726428 | 0.30 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr16_+_18535618 | 0.30 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr8_-_8446668 | 0.30 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr21_-_33995710 | 0.30 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr23_-_27822920 | 0.29 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr6_-_10037207 | 0.29 |
ENSDART00000179701
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr15_-_33807758 | 0.29 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr5_-_22052852 | 0.29 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr13_-_29505604 | 0.29 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr9_-_38399432 | 0.29 |
ENSDART00000148268
|
znf142
|
zinc finger protein 142 |
| chr22_-_16154771 | 0.29 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr25_-_37284370 | 0.28 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr18_+_41560822 | 0.28 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr9_+_23770666 | 0.28 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr11_-_24523161 | 0.28 |
ENSDART00000191936
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr8_-_8446430 | 0.27 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr15_+_26941063 | 0.27 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr12_-_35505610 | 0.27 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr1_-_51038885 | 0.27 |
ENSDART00000035150
|
spast
|
spastin |
| chr21_+_3796620 | 0.26 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr18_+_41561285 | 0.26 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr21_+_22558187 | 0.26 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr23_-_22113455 | 0.26 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr12_-_13966184 | 0.26 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| chr5_+_60919378 | 0.25 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr16_-_32582798 | 0.25 |
ENSDART00000137936
|
fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_-_12477224 | 0.25 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr14_-_33177935 | 0.25 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr18_-_44847855 | 0.24 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr22_-_35063526 | 0.24 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr20_-_29498178 | 0.24 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr18_-_15532016 | 0.24 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr11_+_34523132 | 0.23 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr16_-_53800047 | 0.23 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr17_+_28670132 | 0.23 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr13_+_2442841 | 0.23 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr11_+_30817943 | 0.22 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr4_+_6833583 | 0.22 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr8_+_12930216 | 0.22 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr8_-_31701157 | 0.22 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr12_-_31422433 | 0.21 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr14_+_35024521 | 0.21 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr5_+_40299568 | 0.21 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr22_-_14367966 | 0.20 |
ENSDART00000188796
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr12_+_18358082 | 0.20 |
ENSDART00000123315
|
si:dkey-7e14.3
|
si:dkey-7e14.3 |
| chr3_-_56871330 | 0.20 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr15_+_21712328 | 0.20 |
ENSDART00000192553
|
NKAPD1
|
zgc:162339 |
| chr1_+_33647682 | 0.20 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr11_-_20071642 | 0.20 |
ENSDART00000162931
ENSDART00000159928 ENSDART00000191443 ENSDART00000121722 |
si:dkey-274m17.3
|
si:dkey-274m17.3 |
| chr12_-_10476448 | 0.20 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr17_-_14966384 | 0.20 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr5_-_56924747 | 0.19 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr17_-_1705013 | 0.19 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr11_-_17713987 | 0.19 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr5_-_9931107 | 0.19 |
ENSDART00000077245
|
tor4ab
|
torsin family 4, member Ab |
| chr7_+_38808027 | 0.19 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr13_-_29505946 | 0.18 |
ENSDART00000026679
|
cdhr1a
|
cadherin-related family member 1a |
| chr11_+_25681335 | 0.18 |
ENSDART00000121478
ENSDART00000190877 |
otud5b
|
OTU deubiquitinase 5b |
| chr2_+_25198648 | 0.18 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_+_18612421 | 0.18 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr17_-_50220228 | 0.18 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr3_+_20012891 | 0.18 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr7_+_29080684 | 0.17 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr6_+_52873822 | 0.17 |
ENSDART00000103138
|
or137-3
|
odorant receptor, family H, subfamily 137, member 3 |
| chr6_+_52895008 | 0.17 |
ENSDART00000136143
|
or137-7
|
odorant receptor, family H, subfamily 137, member 7 |
| chr6_+_52903302 | 0.17 |
ENSDART00000143962
|
or137-6
|
odorant receptor, family H, subfamily 137, member 6 |
| chr7_+_24023653 | 0.16 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr6_+_46431848 | 0.16 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr1_+_6817292 | 0.16 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr2_+_47471943 | 0.14 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr2_+_47471647 | 0.14 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr22_-_21755524 | 0.14 |
ENSDART00000149635
|
tle2b
|
transducin like enhancer of split 2b |
| chr3_-_4501026 | 0.14 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr7_+_49800849 | 0.14 |
ENSDART00000164838
ENSDART00000187888 |
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr6_+_52911338 | 0.14 |
ENSDART00000065710
|
si:dkeyp-3f10.14
|
si:dkeyp-3f10.14 |
| chr9_+_34950942 | 0.14 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxi1+foxi2+foxi3a+foxi3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.4 | 1.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.9 | GO:0010259 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.9 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.7 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.5 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.4 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.5 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.5 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.9 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.4 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 1.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 1.0 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 1.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 0.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 0.9 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.4 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.4 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.6 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.4 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 1.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.6 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |