Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
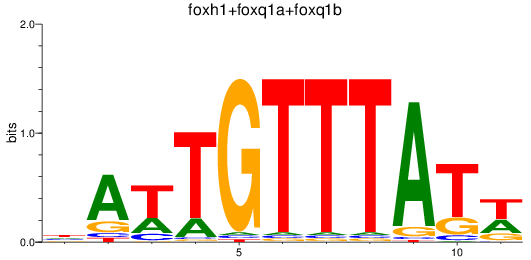
Results for foxh1+foxq1a+foxq1b
Z-value: 0.52
Transcription factors associated with foxh1+foxq1a+foxq1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxq1a
|
ENSDARG00000030896 | forkhead box Q1a |
|
foxq1b
|
ENSDARG00000055395 | forkhead box Q1b |
|
foxh1
|
ENSDARG00000055630 | forkhead box H1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxq1a | dr11_v1_chr2_-_722156_722170 | -0.17 | 6.6e-01 | Click! |
| foxh1 | dr11_v1_chr12_-_13730501_13730501 | 0.14 | 7.1e-01 | Click! |
| foxq1b | dr11_v1_chr20_+_26683933_26683940 | 0.11 | 7.7e-01 | Click! |
Activity profile of foxh1+foxq1a+foxq1b motif
Sorted Z-values of foxh1+foxq1a+foxq1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_19998723 | 0.30 |
ENSDART00000173458
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr7_-_19999152 | 0.27 |
ENSDART00000173881
ENSDART00000100798 |
trip6
|
thyroid hormone receptor interactor 6 |
| chr21_+_4116437 | 0.25 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr11_-_8126223 | 0.23 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr17_+_24590177 | 0.19 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr16_-_13818061 | 0.18 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr4_+_11723852 | 0.16 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr7_+_24153070 | 0.15 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr25_+_2776865 | 0.14 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr12_+_19408373 | 0.14 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr7_-_53117131 | 0.13 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr17_+_15559046 | 0.13 |
ENSDART00000187126
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr6_-_8466717 | 0.13 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr1_+_9290103 | 0.13 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr5_+_8196264 | 0.13 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr22_+_15960514 | 0.13 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr1_+_53321878 | 0.13 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_-_8895130 | 0.13 |
ENSDART00000144673
ENSDART00000138527 |
rab20
|
RAB20, member RAS oncogene family |
| chr5_-_45958838 | 0.13 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr10_-_6587066 | 0.13 |
ENSDART00000171833
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr8_+_8643901 | 0.12 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr14_-_24410673 | 0.12 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr20_-_16156419 | 0.12 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr23_+_17981127 | 0.11 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr7_-_60351876 | 0.11 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr7_-_60351537 | 0.11 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr22_-_29689981 | 0.11 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr5_-_32074534 | 0.11 |
ENSDART00000112342
ENSDART00000135855 |
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr11_+_24348425 | 0.11 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr22_+_15960005 | 0.11 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr6_+_54680730 | 0.11 |
ENSDART00000074602
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr22_+_15959844 | 0.10 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr4_+_75200467 | 0.10 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr13_-_23956178 | 0.10 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr24_+_7861373 | 0.10 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr9_-_27398369 | 0.10 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr23_+_17980875 | 0.10 |
ENSDART00000163452
|
chia.6
|
chitinase, acidic.6 |
| chr13_-_23956361 | 0.10 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr6_+_13787855 | 0.10 |
ENSDART00000182899
|
tmem198b
|
transmembrane protein 198b |
| chr15_-_16183583 | 0.10 |
ENSDART00000062335
|
glod4
|
glyoxalase domain containing 4 |
| chr17_+_28005763 | 0.10 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr25_+_18711804 | 0.09 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr8_+_40476811 | 0.09 |
ENSDART00000129772
|
gck
|
glucokinase (hexokinase 4) |
| chr22_-_5171829 | 0.09 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr9_-_30494727 | 0.09 |
ENSDART00000148729
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr16_+_24733741 | 0.09 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr12_+_5251647 | 0.09 |
ENSDART00000124097
|
plce1
|
phospholipase C, epsilon 1 |
| chr7_+_28724919 | 0.09 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr17_+_44441042 | 0.09 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr1_-_10048514 | 0.09 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr9_+_34397843 | 0.09 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr1_-_23308225 | 0.09 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr5_-_28767573 | 0.09 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr5_+_44944778 | 0.09 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr24_+_39586701 | 0.09 |
ENSDART00000136447
|
si:dkey-161j23.7
|
si:dkey-161j23.7 |
| chr20_-_44055095 | 0.09 |
ENSDART00000125898
ENSDART00000082265 |
runx2b
|
runt-related transcription factor 2b |
| chr12_+_18906939 | 0.09 |
ENSDART00000186074
|
josd1
|
Josephin domain containing 1 |
| chr11_-_27057572 | 0.09 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr22_-_15437242 | 0.09 |
ENSDART00000063719
|
rpp21
|
ribonuclease P 21 subunit |
| chr21_-_44772738 | 0.08 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr12_+_18906407 | 0.08 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr21_+_26733529 | 0.08 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr14_-_15990361 | 0.08 |
ENSDART00000168075
|
trim105
|
tripartite motif containing 105 |
| chr10_+_9595575 | 0.08 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr10_+_20180163 | 0.08 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr12_-_28537615 | 0.08 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr5_+_25762271 | 0.08 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr7_-_17425535 | 0.08 |
ENSDART00000098053
ENSDART00000161873 |
nitr3c
|
novel immune-type receptor 3c |
| chr9_+_20554896 | 0.08 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr5_+_37837245 | 0.08 |
ENSDART00000171617
|
epd
|
ependymin |
| chr13_+_25286538 | 0.08 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr22_+_24559947 | 0.08 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr3_-_43356082 | 0.07 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr15_-_37875601 | 0.07 |
ENSDART00000122439
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr9_+_34397516 | 0.07 |
ENSDART00000011304
ENSDART00000192973 |
med14
|
mediator complex subunit 14 |
| chr9_+_50007556 | 0.07 |
ENSDART00000175587
|
slc38a11
|
solute carrier family 38, member 11 |
| chr23_-_41762956 | 0.07 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr8_+_11425048 | 0.07 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr13_+_39182099 | 0.07 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr15_-_19334448 | 0.07 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr23_+_38957472 | 0.07 |
ENSDART00000193836
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr12_-_31422433 | 0.07 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr16_+_25116827 | 0.07 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr10_+_14488625 | 0.07 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr5_-_33769211 | 0.07 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr17_+_24111392 | 0.07 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr4_+_15011341 | 0.07 |
ENSDART00000124452
|
ube2h
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) |
| chr25_+_36045072 | 0.07 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr24_+_7884880 | 0.07 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr25_-_20024829 | 0.06 |
ENSDART00000140182
ENSDART00000174776 |
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr10_-_8079737 | 0.06 |
ENSDART00000059014
ENSDART00000179549 |
zgc:173443
si:ch211-251f6.6
|
zgc:173443 si:ch211-251f6.6 |
| chr10_+_31646020 | 0.06 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr14_-_38809561 | 0.06 |
ENSDART00000159159
|
sil1
|
SIL1 nucleotide exchange factor |
| chr11_-_25257595 | 0.06 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr18_-_45761868 | 0.06 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr7_+_29080684 | 0.06 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr11_-_25257045 | 0.06 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr1_+_19083501 | 0.06 |
ENSDART00000002886
ENSDART00000131948 |
exosc9
|
exosome component 9 |
| chr12_+_27704015 | 0.06 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr14_+_34495216 | 0.06 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr4_+_23125689 | 0.06 |
ENSDART00000077854
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr24_-_37338162 | 0.06 |
ENSDART00000056303
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr2_-_10631767 | 0.06 |
ENSDART00000190033
|
mtf2
|
metal response element binding transcription factor 2 |
| chr24_+_26329018 | 0.06 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr15_-_7337537 | 0.05 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr11_+_22134540 | 0.05 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr1_+_32054159 | 0.05 |
ENSDART00000181442
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr17_+_25856671 | 0.05 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr2_-_55298075 | 0.05 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr12_-_23365737 | 0.05 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr18_+_25225524 | 0.05 |
ENSDART00000055563
|
si:dkeyp-59c12.1
|
si:dkeyp-59c12.1 |
| chr9_+_8894788 | 0.05 |
ENSDART00000132068
|
naxd
|
NAD(P)HX dehydratase |
| chr3_+_40289418 | 0.05 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr14_-_33177935 | 0.05 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr17_+_34244345 | 0.05 |
ENSDART00000006058
|
eif2s1a
|
eukaryotic translation initiation factor 2, subunit 1 alpha a |
| chr8_-_13210959 | 0.05 |
ENSDART00000142224
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr11_-_24523161 | 0.05 |
ENSDART00000191936
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr17_-_11417551 | 0.05 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr23_+_2191773 | 0.05 |
ENSDART00000190876
ENSDART00000126768 |
cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr15_-_36727462 | 0.05 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr2_+_31957554 | 0.05 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr19_-_48391415 | 0.05 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr1_-_54191626 | 0.05 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr4_-_75157223 | 0.05 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr8_+_25034544 | 0.05 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr2_+_15128418 | 0.05 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr15_-_17169935 | 0.05 |
ENSDART00000110111
|
cul5a
|
cullin 5a |
| chr24_+_7336807 | 0.05 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr8_-_44223899 | 0.05 |
ENSDART00000143020
|
stx2b
|
syntaxin 2b |
| chr24_-_26854032 | 0.05 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr12_-_5455936 | 0.05 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr21_-_13225402 | 0.05 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr7_+_24494299 | 0.05 |
ENSDART00000087568
|
nelfa
|
negative elongation factor complex member A |
| chr1_+_51191049 | 0.05 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr17_-_44440832 | 0.05 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr11_-_22303678 | 0.05 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr5_-_37997774 | 0.05 |
ENSDART00000139616
ENSDART00000167694 |
si:dkey-111e8.1
|
si:dkey-111e8.1 |
| chr16_+_35535375 | 0.04 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr18_+_16246806 | 0.04 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr16_+_9580699 | 0.04 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_+_26676758 | 0.04 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr18_+_13077800 | 0.04 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr8_-_11324143 | 0.04 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr8_-_16592491 | 0.04 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr3_-_24456451 | 0.04 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr19_+_25971000 | 0.04 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr23_+_22819971 | 0.04 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr17_-_11417904 | 0.04 |
ENSDART00000103228
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr15_-_7337148 | 0.04 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr19_+_19756425 | 0.04 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr20_+_35473288 | 0.04 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr3_-_40284744 | 0.04 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr23_-_41762797 | 0.04 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr1_-_45580787 | 0.04 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr17_-_23709347 | 0.04 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr10_+_40321067 | 0.04 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr1_+_19538299 | 0.03 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr7_+_52761841 | 0.03 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr5_+_29726428 | 0.03 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr1_-_53756851 | 0.03 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr10_+_22775253 | 0.03 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr19_+_4139065 | 0.03 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr24_+_37338169 | 0.03 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr12_+_27156943 | 0.03 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr13_+_12396316 | 0.03 |
ENSDART00000143557
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr4_+_25181572 | 0.03 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr17_-_18888959 | 0.03 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr7_+_52950449 | 0.03 |
ENSDART00000187559
|
cdkn2aip
|
CDKN2A interacting protein |
| chr12_-_34716037 | 0.03 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr2_+_7132292 | 0.03 |
ENSDART00000153404
ENSDART00000012119 |
zgc:110366
|
zgc:110366 |
| chr20_-_45062514 | 0.03 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr9_+_34950942 | 0.03 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
| chr12_+_32729470 | 0.03 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr7_+_9290929 | 0.03 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr1_+_35790082 | 0.03 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr12_-_3237561 | 0.03 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr8_-_44223473 | 0.03 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr23_-_43595956 | 0.03 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr7_-_18547420 | 0.03 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr19_+_19777437 | 0.03 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr1_+_46194333 | 0.03 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr8_+_48484455 | 0.03 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr7_+_38808027 | 0.03 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr22_-_29906764 | 0.03 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr9_+_11293830 | 0.03 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr10_+_573667 | 0.02 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr15_+_7992906 | 0.02 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr6_-_6423885 | 0.02 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr5_-_13766651 | 0.02 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr25_-_26893006 | 0.02 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr6_+_33881372 | 0.02 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr5_-_67365750 | 0.02 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr17_+_26803470 | 0.02 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr21_+_10021823 | 0.02 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr3_+_54047342 | 0.02 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr8_-_43847138 | 0.02 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr3_-_49138004 | 0.02 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr8_-_25034411 | 0.02 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr20_-_25748407 | 0.02 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr11_+_24046179 | 0.02 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxh1+foxq1a+foxq1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.0 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |