Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
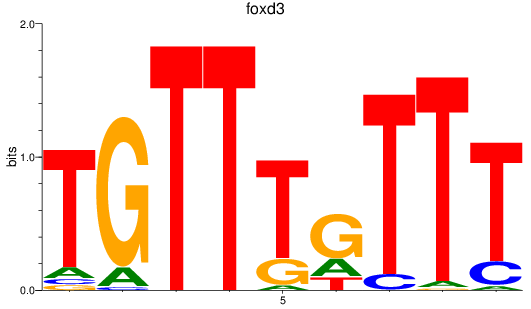
Results for foxd3
Z-value: 2.87
Transcription factors associated with foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd3
|
ENSDARG00000021032 | forkhead box D3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd3 | dr11_v1_chr6_-_32093830_32093830 | -0.78 | 1.2e-02 | Click! |
Activity profile of foxd3 motif
Sorted Z-values of foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_4245902 | 5.21 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr22_+_24559947 | 4.77 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr5_+_57924611 | 4.53 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr5_+_36768674 | 4.48 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr15_+_38299385 | 4.39 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr22_-_10591876 | 4.32 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr17_+_14965570 | 4.07 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr1_-_33647138 | 3.95 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr12_-_22238004 | 3.69 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr15_+_38299563 | 3.57 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr10_-_14929392 | 3.43 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr12_+_28888975 | 3.42 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr16_+_43077909 | 3.11 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr4_-_16628801 | 3.10 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr13_+_37653851 | 3.02 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr7_-_51773166 | 3.02 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr20_-_6532462 | 2.94 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr2_-_38992304 | 2.90 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr19_+_1510971 | 2.88 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr23_+_19590006 | 2.88 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr5_-_9216758 | 2.79 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr1_-_33645967 | 2.78 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr11_+_31864921 | 2.78 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr7_-_48251234 | 2.73 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr16_-_47381519 | 2.63 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr4_-_77116266 | 2.61 |
ENSDART00000174249
|
CU467646.2
|
|
| chr23_+_22873415 | 2.60 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr15_+_19990068 | 2.57 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr8_-_25566347 | 2.53 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr24_+_36317544 | 2.52 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr12_-_28791969 | 2.50 |
ENSDART00000153073
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr22_-_28777557 | 2.48 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr22_+_23359369 | 2.44 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr4_-_77120928 | 2.39 |
ENSDART00000174154
|
CU467646.1
|
|
| chr14_-_897874 | 2.39 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr19_-_34995629 | 2.38 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr20_+_13883131 | 2.37 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr6_+_4255319 | 2.34 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr12_+_20691310 | 2.33 |
ENSDART00000064335
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr5_+_13472234 | 2.30 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr14_+_32926385 | 2.30 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr13_-_35459928 | 2.29 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr2_-_44777592 | 2.29 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr6_-_41135215 | 2.28 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr5_-_3960161 | 2.28 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr12_-_4301234 | 2.27 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr17_-_40956035 | 2.27 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr9_-_52814204 | 2.24 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr8_-_6825588 | 2.24 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr12_-_28794957 | 2.23 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr1_-_6028876 | 2.23 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr8_+_8643901 | 2.20 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr22_+_5574952 | 2.19 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr22_-_22337382 | 2.19 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr13_-_22961605 | 2.18 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr8_+_10862353 | 2.16 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr13_+_31716820 | 2.16 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr19_+_29303847 | 2.14 |
ENSDART00000009149
|
maco1a
|
macoilin 1a |
| chr24_+_10027902 | 2.09 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr3_+_53116172 | 2.08 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr22_-_17671348 | 2.07 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr2_+_34967022 | 2.06 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr11_+_45286911 | 2.04 |
ENSDART00000181763
|
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr3_-_34561624 | 2.01 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr16_+_25259313 | 2.01 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr8_+_37749263 | 2.00 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr19_-_6193448 | 2.00 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr8_+_42917515 | 1.98 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr15_-_16076399 | 1.96 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr9_+_2762270 | 1.96 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr11_-_270210 | 1.96 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr3_+_46271911 | 1.94 |
ENSDART00000186557
ENSDART00000113531 |
mkl2b
|
MKL/myocardin-like 2b |
| chr3_-_43650189 | 1.93 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr3_-_29910547 | 1.93 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr18_+_17534627 | 1.92 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr22_-_25417132 | 1.92 |
ENSDART00000099060
|
zgc:171781
|
zgc:171781 |
| chr7_+_15736230 | 1.90 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr17_-_2685026 | 1.90 |
ENSDART00000191014
ENSDART00000179309 |
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr20_+_13883353 | 1.89 |
ENSDART00000188006
|
nek2
|
NIMA-related kinase 2 |
| chr21_+_3960583 | 1.89 |
ENSDART00000149788
|
setx
|
senataxin |
| chr25_+_28158352 | 1.88 |
ENSDART00000151854
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr19_+_31532043 | 1.87 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr5_-_26247973 | 1.87 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr3_-_2623176 | 1.86 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr21_+_4116437 | 1.86 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr25_+_16214854 | 1.85 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr9_+_38458193 | 1.85 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr17_+_24111392 | 1.85 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr2_+_1988036 | 1.85 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr5_-_49951106 | 1.84 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr12_-_4835378 | 1.84 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr3_-_60711127 | 1.84 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr16_+_47207691 | 1.83 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr6_+_2174082 | 1.83 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr5_-_38384289 | 1.83 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr7_-_48263516 | 1.82 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr5_+_61361815 | 1.81 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr19_-_2822372 | 1.80 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr12_+_20412293 | 1.78 |
ENSDART00000115015
ENSDART00000177573 |
arhgap17a
|
Rho GTPase activating protein 17a |
| chr8_+_42941555 | 1.77 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr17_-_2690083 | 1.76 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr17_+_13664442 | 1.76 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr12_-_17863467 | 1.75 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr5_-_30074332 | 1.75 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr5_-_23485161 | 1.73 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr16_-_49646625 | 1.73 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr23_-_18707418 | 1.73 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr5_-_57289872 | 1.72 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr5_+_66250856 | 1.71 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr5_-_29122834 | 1.71 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr7_+_13830052 | 1.70 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr4_-_5691257 | 1.70 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr2_+_44512324 | 1.69 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr7_+_13824150 | 1.67 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr18_+_15271993 | 1.67 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr17_-_29311835 | 1.67 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr19_+_31585341 | 1.67 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr11_+_18175893 | 1.65 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr18_+_25546227 | 1.65 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr6_-_29377092 | 1.65 |
ENSDART00000078665
|
tmem131
|
transmembrane protein 131 |
| chr15_-_7337537 | 1.63 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr1_+_53321878 | 1.63 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr10_+_15603082 | 1.62 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr1_+_29211540 | 1.62 |
ENSDART00000053924
|
irs2a
|
insulin receptor substrate 2a |
| chr17_-_43594864 | 1.62 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr15_-_17099560 | 1.61 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr5_-_16475374 | 1.60 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_-_16475682 | 1.60 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr2_+_44518636 | 1.60 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr5_-_3991655 | 1.59 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr11_+_18183220 | 1.58 |
ENSDART00000113468
|
LO018315.10
|
|
| chr16_-_35532937 | 1.58 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr23_+_38171186 | 1.58 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr11_-_24523161 | 1.57 |
ENSDART00000191936
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr21_+_15723069 | 1.57 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr16_-_41762983 | 1.57 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr20_-_4793450 | 1.56 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr25_+_2776865 | 1.55 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr5_-_31773208 | 1.55 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr16_+_25608778 | 1.54 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr5_-_23574234 | 1.53 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_+_6833735 | 1.53 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr3_-_15119856 | 1.52 |
ENSDART00000138328
|
xpo6
|
exportin 6 |
| chr7_+_52950449 | 1.52 |
ENSDART00000187559
|
cdkn2aip
|
CDKN2A interacting protein |
| chr14_-_34633960 | 1.52 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr4_-_20135406 | 1.52 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr6_+_40922572 | 1.52 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_+_45139196 | 1.52 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_26432677 | 1.51 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr6_-_40922971 | 1.51 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr13_-_18011168 | 1.51 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr19_+_41479990 | 1.51 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr16_-_41488023 | 1.50 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr18_+_27077853 | 1.49 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr22_-_28777374 | 1.49 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr5_+_3891485 | 1.49 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr2_-_57110477 | 1.48 |
ENSDART00000181132
|
slc25a42
|
solute carrier family 25, member 42 |
| chr15_+_23657051 | 1.48 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr6_-_25201810 | 1.47 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr7_-_9674073 | 1.46 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr4_+_14981854 | 1.46 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr8_-_49304602 | 1.45 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr13_+_45582391 | 1.44 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr10_-_14929630 | 1.44 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr15_-_7337148 | 1.44 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr13_-_11378355 | 1.44 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr7_+_52712807 | 1.43 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr25_-_13789955 | 1.43 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr13_+_50778187 | 1.42 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr15_+_26940569 | 1.42 |
ENSDART00000189636
ENSDART00000077172 |
bcas3
|
breast carcinoma amplified sequence 3 |
| chr22_-_3299100 | 1.42 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr10_-_33251876 | 1.42 |
ENSDART00000184565
|
bcl7ba
|
BCL tumor suppressor 7Ba |
| chr16_+_38820486 | 1.42 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr5_-_23517747 | 1.42 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| chr3_+_32577994 | 1.41 |
ENSDART00000180643
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr8_+_13368150 | 1.40 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr23_-_33738570 | 1.40 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr23_-_22113455 | 1.40 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr5_+_25311309 | 1.40 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr4_+_9467049 | 1.39 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr16_-_41487589 | 1.39 |
ENSDART00000188115
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr3_-_26191960 | 1.39 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr15_-_41689684 | 1.39 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr21_-_13856689 | 1.38 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr12_-_7234915 | 1.38 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr7_-_40630698 | 1.38 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr1_+_45707219 | 1.38 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr1_+_51191049 | 1.37 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr8_+_30742898 | 1.37 |
ENSDART00000018475
|
snrpd3
|
small nuclear ribonucleoprotein D3 polypeptide |
| chr3_-_55537096 | 1.37 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr1_+_44173245 | 1.37 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_-_40519340 | 1.36 |
ENSDART00000114659
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr1_-_26675969 | 1.36 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
| chr14_+_35428152 | 1.36 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr11_+_45287541 | 1.36 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr15_+_29393519 | 1.35 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr21_+_5960443 | 1.35 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr6_+_12527725 | 1.34 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr13_+_11828516 | 1.34 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr5_+_29652513 | 1.34 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.2 | 3.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 1.0 | 5.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 1.0 | 3.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.8 | 4.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.8 | 3.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.8 | 2.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.8 | 3.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.8 | 3.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.7 | 0.7 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.7 | 2.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.7 | 2.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.7 | 3.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.6 | 3.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.6 | 2.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 1.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.5 | 2.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.5 | 1.6 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.5 | 1.6 | GO:0060281 | regulation of oocyte development(GO:0060281) regulation of oocyte maturation(GO:1900193) regulation of cell maturation(GO:1903429) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.5 | 2.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.5 | 2.0 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.5 | 3.4 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 1.9 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 2.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.4 | 3.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.4 | 3.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 4.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 2.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.4 | 1.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.4 | 1.3 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 | 1.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.4 | 1.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 1.2 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.4 | 1.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 1.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.4 | 3.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.4 | 1.5 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.4 | 1.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 1.1 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.4 | 3.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 1.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 1.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.4 | 1.8 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.4 | 1.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.4 | 4.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 1.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 1.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 3.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 1.3 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 1.0 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.3 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 3.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 1.3 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.3 | 1.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.3 | 1.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 1.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.3 | 1.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 1.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 1.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 1.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 2.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 0.8 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 1.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 0.8 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 1.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 0.8 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.3 | 3.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 1.0 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 2.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.7 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 0.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 0.9 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 2.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.8 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.2 | 1.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 2.0 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.2 | 1.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 1.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 2.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 1.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 2.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 2.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 3.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 1.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.2 | 1.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.6 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 | 1.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 2.2 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.2 | 2.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 2.7 | GO:0010575 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.2 | 1.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.6 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 2.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 1.1 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.2 | 1.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 1.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 2.2 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.2 | 1.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.8 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.0 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.2 | 1.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.5 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 3.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.9 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 0.8 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 0.6 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 0.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 3.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.4 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.6 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.7 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.7 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 2.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.6 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 1.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 1.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 2.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.6 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 2.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.1 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 3.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 1.2 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 1.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.1 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 4.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.7 | GO:1900117 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 1.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.5 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 2.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 1.0 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.2 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.1 | 0.7 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 4.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 2.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.3 | GO:0039531 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 2.6 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 1.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.9 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.5 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:0045110 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.8 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.1 | 2.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.3 | GO:2000223 | posterior lateral line neuromast deposition(GO:0048922) regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.0 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.2 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.6 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 3.2 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 2.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 1.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 1.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.3 | GO:0001909 | leukocyte mediated cytotoxicity(GO:0001909) |
| 0.1 | 7.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 2.6 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte migration(GO:0002686) negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.6 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.8 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.2 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.1 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 5.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 1.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.6 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 5.8 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 1.2 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.2 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 3.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.1 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.6 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.4 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 2.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.1 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.1 | 1.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.7 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 0.5 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.0 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.2 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.6 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.1 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.8 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 5.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.5 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 1.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.0 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.8 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 5.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.4 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.0 | 0.8 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.7 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.4 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 1.2 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 2.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.7 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 2.4 | GO:0034249 | negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 4.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 1.3 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.7 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.9 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 2.5 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.6 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.8 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.1 | GO:0048278 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 1.0 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 2.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.2 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.5 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 1.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.5 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.7 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 9.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.0 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 2.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 3.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 2.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.4 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.5 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.0 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.0 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.1 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.8 | 3.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.7 | 3.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.6 | 3.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.5 | 2.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.5 | 2.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 5.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 1.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.3 | 2.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 1.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 2.7 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.3 | 2.1 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.3 | 0.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 5.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 3.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.3 | 1.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 1.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 1.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 0.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 3.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 3.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 3.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 1.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 0.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 2.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 6.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.9 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.7 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.2 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.4 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 8.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 6.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.0 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 11.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 5.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 5.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 4.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 9.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 8.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 8.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 7.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 3.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 5.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 10.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.7 | 3.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.7 | 3.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.7 | 2.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.6 | 1.9 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.6 | 1.9 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.6 | 1.7 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.6 | 3.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.5 | 2.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 1.5 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.5 | 4.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.5 | 1.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 3.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.4 | 3.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.4 | 3.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 2.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 3.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 1.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 2.6 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.4 | 1.4 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 1.3 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.2 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.3 | 2.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 5.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 2.7 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 1.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.3 | 1.0 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 0.8 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 2.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.4 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.2 | 0.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 4.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 5.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 3.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 2.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 3.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.6 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 2.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 2.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 4.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 2.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.6 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.7 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 2.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.8 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.4 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 2.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 4.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.8 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 1.2 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 1.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.4 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 1.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.4 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 2.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.7 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 12.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.2 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 2.0 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 9.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 15.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 8.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 2.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 6.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.9 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 3.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 7.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.8 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 2.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 10.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 11.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 3.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 3.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 3.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 4.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 8.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 0.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 2.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.8 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.1 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |