Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
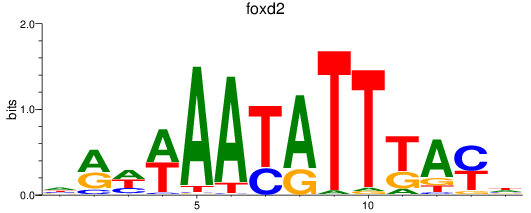
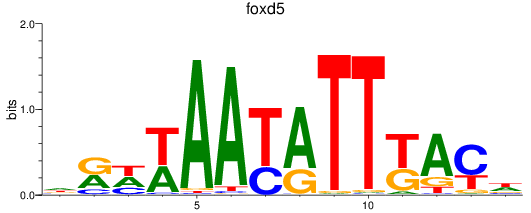
Results for foxd2_foxd5
Z-value: 1.30
Transcription factors associated with foxd2_foxd5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd2
|
ENSDARG00000058133 | forkhead box D2 |
|
foxd5
|
ENSDARG00000042485 | forkhead box D5 |
|
foxd5
|
ENSDARG00000109712 | forkhead box D5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd2 | dr11_v1_chr8_+_19674369_19674369 | 0.84 | 5.0e-03 | Click! |
| foxd5 | dr11_v1_chr8_+_30452945_30452945 | 0.28 | 4.7e-01 | Click! |
Activity profile of foxd2_foxd5 motif
Sorted Z-values of foxd2_foxd5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12013043 | 3.54 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr14_+_21107032 | 2.91 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr22_-_17595310 | 2.76 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr14_+_11458044 | 2.66 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr14_+_11457500 | 2.58 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr8_-_16697615 | 2.58 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr3_+_36424055 | 2.46 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr6_+_2097690 | 2.44 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr16_+_11724230 | 2.41 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr8_+_16004551 | 2.26 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_+_8660158 | 2.15 |
ENSDART00000169141
|
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr17_+_21964472 | 2.11 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr1_-_30473422 | 2.08 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr18_-_46354269 | 2.05 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr22_-_23668356 | 2.01 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr10_+_36650222 | 1.84 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr25_+_29160102 | 1.84 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr5_-_14344647 | 1.80 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr8_+_16004154 | 1.77 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_+_32206378 | 1.71 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr22_-_11438627 | 1.70 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr12_-_30583668 | 1.70 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr6_-_8264751 | 1.63 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr4_-_16412084 | 1.62 |
ENSDART00000188460
|
dcn
|
decorin |
| chr13_+_32144370 | 1.61 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr5_+_26795465 | 1.59 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr22_+_11756040 | 1.57 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr17_-_10838434 | 1.54 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr23_-_4925641 | 1.46 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr14_-_4121052 | 1.45 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr1_-_35924495 | 1.42 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr25_+_19149241 | 1.37 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr23_-_44219902 | 1.36 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr21_+_44684577 | 1.33 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr9_+_34127005 | 1.30 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr8_-_13013123 | 1.26 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr4_-_25215968 | 1.25 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr3_-_50863370 | 1.24 |
ENSDART00000169771
ENSDART00000165083 |
pmp22a
|
peripheral myelin protein 22a |
| chr18_-_6803424 | 1.19 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr19_-_22387141 | 1.19 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr5_+_67662430 | 1.19 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr24_+_22485710 | 1.16 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr2_-_30693742 | 1.16 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr23_-_26077038 | 1.14 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr20_-_27225876 | 1.14 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr22_+_5663529 | 1.13 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr17_-_10073926 | 1.11 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr20_+_36233873 | 1.05 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr13_-_30662403 | 1.03 |
ENSDART00000012457
|
si:dkey-275b16.2
|
si:dkey-275b16.2 |
| chr13_+_32446169 | 0.99 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr21_+_35215810 | 0.97 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr21_+_27416284 | 0.95 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr13_+_29773153 | 0.94 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr2_-_9748039 | 0.88 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr5_-_51756210 | 0.88 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr24_+_7631797 | 0.87 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr6_-_55864687 | 0.87 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr4_-_12723585 | 0.87 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr2_-_37956768 | 0.86 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr8_-_15150131 | 0.85 |
ENSDART00000138253
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr14_-_40389699 | 0.85 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr12_-_34887943 | 0.83 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr23_-_20258490 | 0.83 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr15_+_23911655 | 0.82 |
ENSDART00000156304
|
si:ch1073-145m9.1
|
si:ch1073-145m9.1 |
| chr21_+_17110598 | 0.82 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr19_+_11984725 | 0.81 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr9_+_30108641 | 0.78 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr7_+_65387374 | 0.76 |
ENSDART00000188680
|
CLEC3A
|
C-type lectin domain family 3 member A |
| chr14_-_33894915 | 0.75 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr14_-_4120636 | 0.74 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr4_-_7212875 | 0.74 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr19_-_31402429 | 0.73 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr16_-_35952789 | 0.73 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr2_+_47906240 | 0.73 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr3_-_50139860 | 0.70 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr4_-_21851473 | 0.70 |
ENSDART00000019748
|
lin7a
|
lin-7 homolog A (C. elegans) |
| chr15_+_28368644 | 0.70 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr16_-_38629208 | 0.70 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr14_-_26377044 | 0.66 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr16_-_7228276 | 0.66 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr22_-_13042992 | 0.66 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr24_-_35707552 | 0.65 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr20_-_9963713 | 0.65 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr4_+_12013642 | 0.65 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr5_-_11943750 | 0.64 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr6_-_32703317 | 0.64 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr15_+_24691088 | 0.63 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr20_+_26881600 | 0.63 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr5_+_43530388 | 0.63 |
ENSDART00000190254
ENSDART00000097618 ENSDART00000133006 |
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr2_+_36898982 | 0.63 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr1_-_37087966 | 0.62 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr15_+_28368823 | 0.62 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_11867639 | 0.61 |
ENSDART00000144677
|
mras
|
muscle RAS oncogene homolog |
| chr16_-_44349845 | 0.60 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr23_-_16960972 | 0.59 |
ENSDART00000142546
|
cdc42l2
|
cell division cycle 42 like 2 |
| chr14_-_1635296 | 0.59 |
ENSDART00000186658
|
LO018564.1
|
|
| chr14_+_46410766 | 0.59 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr2_-_5135125 | 0.58 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr19_+_32856907 | 0.57 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr3_-_28828242 | 0.57 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr12_+_23424108 | 0.57 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr12_-_36260532 | 0.55 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr7_+_32722227 | 0.55 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr17_+_8292892 | 0.54 |
ENSDART00000125728
|
si:ch211-236p5.3
|
si:ch211-236p5.3 |
| chr9_-_23147026 | 0.53 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr10_-_8294965 | 0.53 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr18_+_19974289 | 0.53 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr17_-_42213285 | 0.52 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr18_-_24988645 | 0.51 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr2_-_985417 | 0.51 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr3_+_41917499 | 0.51 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_-_31522625 | 0.51 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr16_+_38119004 | 0.50 |
ENSDART00000132087
|
pogzb
|
pogo transposable element derived with ZNF domain b |
| chr5_-_67750907 | 0.49 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr1_-_656693 | 0.49 |
ENSDART00000170483
ENSDART00000166786 |
appa
|
amyloid beta (A4) precursor protein a |
| chr5_+_22970617 | 0.48 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr3_-_21280373 | 0.48 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr6_-_19035749 | 0.47 |
ENSDART00000187714
|
sept9b
|
septin 9b |
| chr17_-_37052622 | 0.47 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr5_+_29671681 | 0.46 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr6_-_40098641 | 0.45 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr19_+_43359075 | 0.45 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr15_-_28677725 | 0.44 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr20_+_34717403 | 0.44 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr9_-_21231297 | 0.44 |
ENSDART00000162578
|
pla1a
|
phospholipase A1 member A |
| chr3_-_16289826 | 0.43 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr16_+_46492994 | 0.43 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr7_+_33314925 | 0.42 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr4_+_7841627 | 0.42 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr5_-_22027357 | 0.42 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr10_-_8295294 | 0.42 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr20_+_2950005 | 0.41 |
ENSDART00000135919
|
amd1
|
adenosylmethionine decarboxylase 1 |
| chr15_-_16121496 | 0.41 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr8_+_32747612 | 0.39 |
ENSDART00000142824
|
hmcn2
|
hemicentin 2 |
| chr3_+_34683096 | 0.38 |
ENSDART00000084432
|
dusp3b
|
dual specificity phosphatase 3b |
| chr22_-_6048054 | 0.38 |
ENSDART00000183239
|
CU468041.2
|
|
| chr16_+_43139504 | 0.38 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr24_-_22756508 | 0.37 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr20_-_25436389 | 0.36 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr16_+_21330634 | 0.36 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr10_-_35236949 | 0.35 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr15_+_32867420 | 0.34 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr20_+_27647436 | 0.34 |
ENSDART00000180557
ENSDART00000185628 |
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr6_+_24661736 | 0.34 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr7_+_56703254 | 0.34 |
ENSDART00000184547
ENSDART00000004964 ENSDART00000147259 ENSDART00000134173 |
nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr17_-_42213822 | 0.33 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr18_-_16181952 | 0.33 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr25_+_24250247 | 0.33 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr18_+_19419120 | 0.32 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr6_+_54187643 | 0.31 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr13_+_32740509 | 0.31 |
ENSDART00000076423
ENSDART00000160138 |
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr8_-_16499508 | 0.30 |
ENSDART00000108659
|
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr5_+_53009083 | 0.30 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr5_-_29671586 | 0.30 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr21_-_32284532 | 0.30 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr19_+_816208 | 0.30 |
ENSDART00000093304
|
nrm
|
nurim |
| chr15_+_45563491 | 0.29 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr18_+_13837746 | 0.29 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr5_-_61624693 | 0.29 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr22_-_9728208 | 0.29 |
ENSDART00000185962
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr18_-_5111449 | 0.28 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr3_-_29506960 | 0.28 |
ENSDART00000141720
|
cyth4a
|
cytohesin 4a |
| chr8_+_26410197 | 0.28 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr3_+_25216790 | 0.28 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr21_-_39327223 | 0.27 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr8_-_44586981 | 0.27 |
ENSDART00000026831
ENSDART00000113945 |
rsph14
|
radial spoke head 14 homolog |
| chr20_+_43083745 | 0.27 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr14_+_36628131 | 0.27 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr3_+_39566999 | 0.26 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr10_+_34315719 | 0.26 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr5_-_3684402 | 0.25 |
ENSDART00000184354
|
FO704648.1
|
|
| chr13_-_18195942 | 0.25 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr13_-_4707018 | 0.25 |
ENSDART00000128422
|
oit3
|
oncoprotein induced transcript 3 |
| chr20_-_51355465 | 0.25 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr22_-_15562933 | 0.25 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr11_+_41858807 | 0.23 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr5_+_13385837 | 0.23 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr18_+_36664654 | 0.22 |
ENSDART00000128707
ENSDART00000098972 |
capn12
|
calpain 12 |
| chr14_+_34966598 | 0.22 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr17_-_24564674 | 0.22 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr16_-_19373310 | 0.21 |
ENSDART00000148294
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr12_-_20120702 | 0.21 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr19_+_46222428 | 0.20 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr8_-_46457233 | 0.20 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr6_+_9870192 | 0.19 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr5_+_54400971 | 0.19 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr12_-_22524388 | 0.18 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr2_-_37960688 | 0.18 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr15_+_24212847 | 0.18 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr16_-_29334672 | 0.18 |
ENSDART00000162835
|
bcan
|
brevican |
| chr24_+_5840258 | 0.18 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr1_+_33697170 | 0.18 |
ENSDART00000131664
|
NSUN3
|
NOP2/Sun RNA methyltransferase family member 3 |
| chr6_-_40697585 | 0.18 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr15_+_45563656 | 0.17 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr8_+_23142946 | 0.17 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr25_+_4760489 | 0.17 |
ENSDART00000167399
|
FP245455.1
|
|
| chr11_+_14284866 | 0.17 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr22_-_15280638 | 0.17 |
ENSDART00000063008
|
mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_40671336 | 0.17 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr5_+_35398745 | 0.16 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd2_foxd5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.6 | 2.6 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.5 | 1.6 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.5 | 1.6 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.4 | 1.8 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 1.5 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 0.8 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 0.8 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 1.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.6 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 3.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.5 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.9 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 2.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 1.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.4 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.3 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.4 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 1.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.8 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 2.6 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.0 | 1.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.3 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.9 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 2.9 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 3.9 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 1.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.9 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.7 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 1.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 4.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.6 | 2.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 1.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 3.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 0.9 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.3 | 1.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.3 | 1.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.9 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.2 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.6 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |