Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
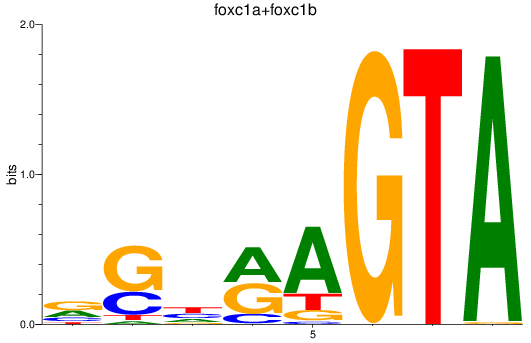
Results for foxc1a+foxc1b
Z-value: 0.80
Transcription factors associated with foxc1a+foxc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxc1b
|
ENSDARG00000055398 | forkhead box C1b |
|
foxc1a
|
ENSDARG00000091481 | forkhead box C1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxc1b | dr11_v1_chr20_+_26702377_26702377 | 0.48 | 1.9e-01 | Click! |
| foxc1a | dr11_v1_chr2_-_689047_689047 | 0.45 | 2.3e-01 | Click! |
Activity profile of foxc1a+foxc1b motif
Sorted Z-values of foxc1a+foxc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_46310204 | 1.22 |
ENSDART00000188713
ENSDART00000184930 |
crybb1l1
|
crystallin, beta B1, like 1 |
| chr25_+_20216159 | 1.14 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr22_+_20720808 | 0.97 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr13_+_28417297 | 0.93 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr11_+_30729745 | 0.85 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr1_+_7540978 | 0.79 |
ENSDART00000147770
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr11_+_11200550 | 0.79 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr1_-_25679339 | 0.76 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr21_+_6556635 | 0.76 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr12_-_26064480 | 0.70 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr12_-_26064105 | 0.69 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr7_+_23907692 | 0.68 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr22_+_5103349 | 0.66 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr2_-_24554416 | 0.57 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr1_-_54947592 | 0.52 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr18_+_15706160 | 0.48 |
ENSDART00000131524
|
si:ch211-264e16.1
|
si:ch211-264e16.1 |
| chr21_-_43606502 | 0.46 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr16_-_42004544 | 0.45 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr24_-_32408404 | 0.45 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr25_+_16601839 | 0.45 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr2_-_50372467 | 0.43 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr5_+_39087364 | 0.43 |
ENSDART00000004286
|
anxa3a
|
annexin A3a |
| chr15_-_20468302 | 0.42 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr15_+_47903864 | 0.41 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr18_+_808911 | 0.40 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr12_-_31794908 | 0.37 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_-_19339285 | 0.37 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr1_-_23274393 | 0.37 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr7_-_43787616 | 0.35 |
ENSDART00000179758
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_+_44911405 | 0.35 |
ENSDART00000182465
|
wu:fc21g02
|
wu:fc21g02 |
| chr2_-_31614277 | 0.34 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr21_-_4032650 | 0.32 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr16_+_7662609 | 0.32 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr6_+_40629066 | 0.32 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr19_-_3381422 | 0.32 |
ENSDART00000105146
|
edn1
|
endothelin 1 |
| chr5_-_65000312 | 0.32 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr8_+_19514294 | 0.31 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr16_-_5721386 | 0.30 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr17_+_53250802 | 0.30 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr9_+_17309195 | 0.29 |
ENSDART00000048548
|
scel
|
sciellin |
| chr20_-_27311675 | 0.29 |
ENSDART00000026088
ENSDART00000148361 |
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr24_-_40726073 | 0.29 |
ENSDART00000168100
|
smyhc2
|
slow myosin heavy chain 2 |
| chr11_+_583142 | 0.29 |
ENSDART00000168157
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr1_-_14332283 | 0.28 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr2_-_43635777 | 0.28 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr20_-_35578435 | 0.27 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr23_+_18944388 | 0.27 |
ENSDART00000104487
|
cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr4_-_4387012 | 0.27 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr11_-_29946927 | 0.27 |
ENSDART00000165182
|
zgc:113276
|
zgc:113276 |
| chr9_-_21918963 | 0.26 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr5_-_28029558 | 0.26 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr21_-_22114625 | 0.26 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr6_-_39275793 | 0.26 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr1_-_45177373 | 0.26 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr15_+_15456910 | 0.25 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr7_-_18691637 | 0.25 |
ENSDART00000183715
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr17_-_12336987 | 0.25 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_-_10828880 | 0.25 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr1_+_35862550 | 0.24 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr11_-_11331052 | 0.24 |
ENSDART00000081765
ENSDART00000160247 |
mrpl4
|
mitochondrial ribosomal protein L4 |
| chr15_+_638457 | 0.23 |
ENSDART00000157162
|
znf1013
|
zinc finger protein 1013 |
| chr8_+_42998944 | 0.23 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr20_+_10723292 | 0.23 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr22_-_10486477 | 0.22 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr12_+_2399130 | 0.21 |
ENSDART00000075179
|
vstm4b
|
V-set and transmembrane domain containing 4b |
| chr6_-_19035749 | 0.21 |
ENSDART00000187714
|
sept9b
|
septin 9b |
| chr21_-_22928214 | 0.21 |
ENSDART00000182760
|
dub
|
duboraya |
| chr17_-_17764801 | 0.21 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr14_-_2602445 | 0.20 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr19_-_26736336 | 0.19 |
ENSDART00000109258
ENSDART00000182802 |
csnk2b
|
casein kinase 2, beta polypeptide |
| chr3_-_18030938 | 0.19 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr9_-_7089303 | 0.19 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr11_+_37250839 | 0.19 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr22_-_10487490 | 0.19 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr11_-_45138857 | 0.18 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr22_-_23000815 | 0.18 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr22_-_26175237 | 0.18 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr6_-_34141935 | 0.18 |
ENSDART00000127063
|
fam183a
|
family with sequence similarity 183, member A |
| chr24_+_9372292 | 0.17 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr14_-_44773864 | 0.17 |
ENSDART00000158386
|
si:dkey-109l4.3
|
si:dkey-109l4.3 |
| chr21_-_30111134 | 0.17 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr6_+_52853384 | 0.17 |
ENSDART00000174155
ENSDART00000164391 |
BX957318.1
|
|
| chr20_-_19365875 | 0.17 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr3_-_50954607 | 0.17 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr21_+_11105007 | 0.17 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr5_-_54300725 | 0.16 |
ENSDART00000163048
|
slc25a35
|
solute carrier family 25, member 35 |
| chr2_+_37804235 | 0.16 |
ENSDART00000076321
|
ltb4r2b
|
leukotriene B4 receptor 2b |
| chr5_-_4219741 | 0.16 |
ENSDART00000100093
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr9_-_24244383 | 0.16 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr25_-_31953516 | 0.15 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr11_-_36046741 | 0.15 |
ENSDART00000184229
|
brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr10_+_15107886 | 0.15 |
ENSDART00000188047
ENSDART00000164095 |
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr14_+_16937997 | 0.15 |
ENSDART00000163013
ENSDART00000167856 |
limch1b
|
LIM and calponin homology domains 1b |
| chr6_-_34008827 | 0.15 |
ENSDART00000191183
ENSDART00000003701 ENSDART00000192502 |
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr7_-_2004159 | 0.15 |
ENSDART00000181965
ENSDART00000173821 |
zgc:171424
zgc:171424
|
zgc:171424 zgc:171424 |
| chr2_-_9816492 | 0.15 |
ENSDART00000138472
ENSDART00000124051 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr7_+_19482877 | 0.15 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr4_-_25064510 | 0.15 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr17_-_53439866 | 0.15 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr25_+_31958911 | 0.14 |
ENSDART00000191394
ENSDART00000090727 ENSDART00000185893 |
duox
|
dual oxidase |
| chr13_-_47800342 | 0.14 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr24_-_21588914 | 0.14 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr4_-_53370 | 0.14 |
ENSDART00000180254
ENSDART00000186529 |
CU856344.2
|
|
| chr16_-_31974482 | 0.14 |
ENSDART00000189073
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr1_+_31638274 | 0.14 |
ENSDART00000057885
|
fgf8b
|
fibroblast growth factor 8 b |
| chr23_-_36316352 | 0.13 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr16_+_38119004 | 0.13 |
ENSDART00000132087
|
pogzb
|
pogo transposable element derived with ZNF domain b |
| chr15_-_18574716 | 0.13 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr1_-_8101495 | 0.13 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr19_-_10730488 | 0.13 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr7_+_19483277 | 0.13 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr25_+_3525494 | 0.12 |
ENSDART00000169775
|
RAB19 (1 of many)
|
RAB19, member RAS oncogene family |
| chr1_-_17663108 | 0.12 |
ENSDART00000131559
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr19_-_17526735 | 0.12 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr1_+_18550864 | 0.12 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr23_+_6272638 | 0.12 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr13_+_31002658 | 0.12 |
ENSDART00000140257
|
lrrc18a
|
leucine rich repeat containing 18a |
| chr9_-_48736388 | 0.12 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr8_-_46525092 | 0.12 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr5_-_12687357 | 0.12 |
ENSDART00000193206
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr14_-_29858883 | 0.12 |
ENSDART00000141034
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr9_-_3866707 | 0.11 |
ENSDART00000010378
|
myo3b
|
myosin IIIB |
| chr23_+_24926407 | 0.11 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr21_-_2310064 | 0.11 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr22_-_8860869 | 0.11 |
ENSDART00000166188
|
CABZ01046427.2
|
|
| chr3_+_12784460 | 0.10 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr16_-_12723324 | 0.10 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr15_-_12319065 | 0.10 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr1_-_9109699 | 0.10 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr12_+_695619 | 0.10 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr9_+_13985567 | 0.10 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr10_+_38512270 | 0.10 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr1_-_45146834 | 0.10 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr2_+_6181383 | 0.10 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr22_-_20289948 | 0.10 |
ENSDART00000132951
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr17_-_50430817 | 0.10 |
ENSDART00000154007
|
si:ch211-235i11.4
|
si:ch211-235i11.4 |
| chr17_-_23412705 | 0.09 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr10_+_23520952 | 0.09 |
ENSDART00000125317
|
zmp:0000000937
|
zmp:0000000937 |
| chr5_-_61942482 | 0.09 |
ENSDART00000097338
|
napaa
|
N-ethylmaleimide-sensitive factor attachment protein, alpha a |
| chr12_-_9438227 | 0.09 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr7_-_22132265 | 0.09 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr5_+_57714902 | 0.08 |
ENSDART00000182860
|
ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr18_+_44080948 | 0.08 |
ENSDART00000191036
|
BX908388.2
|
|
| chr6_-_8480815 | 0.08 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr1_-_37383741 | 0.08 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr21_-_17290941 | 0.08 |
ENSDART00000147993
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr12_-_35095414 | 0.08 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr12_+_20641471 | 0.08 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr4_+_57093908 | 0.08 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr1_-_22338521 | 0.08 |
ENSDART00000176849
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr14_-_22390821 | 0.07 |
ENSDART00000054396
|
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr16_-_4203022 | 0.07 |
ENSDART00000018686
|
rrp15
|
ribosomal RNA processing 15 homolog |
| chr20_+_51479263 | 0.07 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr18_+_25752592 | 0.07 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr8_-_15398760 | 0.07 |
ENSDART00000162011
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr18_-_16792561 | 0.07 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr9_-_23994225 | 0.07 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr23_+_16910071 | 0.07 |
ENSDART00000104793
|
si:dkey-147f3.8
|
si:dkey-147f3.8 |
| chr5_+_23597144 | 0.07 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr4_-_14899113 | 0.06 |
ENSDART00000137829
|
si:dkey-14k9.3
|
si:dkey-14k9.3 |
| chr3_+_46763745 | 0.06 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr15_-_3976035 | 0.06 |
ENSDART00000168061
|
si:ch73-309g22.1
|
si:ch73-309g22.1 |
| chr10_+_17714866 | 0.06 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr5_+_37890521 | 0.06 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr1_-_37383539 | 0.06 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr16_-_24815091 | 0.06 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_40158146 | 0.05 |
ENSDART00000145694
|
si:ch211-113e8.9
|
si:ch211-113e8.9 |
| chr12_-_37941733 | 0.05 |
ENSDART00000130167
|
CABZ01046949.1
|
|
| chr25_+_32474031 | 0.05 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr8_+_32747612 | 0.05 |
ENSDART00000142824
|
hmcn2
|
hemicentin 2 |
| chr24_-_12958668 | 0.05 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr22_+_26793389 | 0.05 |
ENSDART00000165381
|
pimr69
|
Pim proto-oncogene, serine/threonine kinase, related 69 |
| chr17_-_33415740 | 0.04 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr2_-_36914281 | 0.04 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr8_-_36370552 | 0.04 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr5_-_50640171 | 0.04 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr23_-_42752387 | 0.04 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr7_+_49695904 | 0.04 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr2_-_51500957 | 0.04 |
ENSDART00000172481
|
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr15_-_36347858 | 0.04 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr15_-_6966221 | 0.04 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr8_-_48770163 | 0.04 |
ENSDART00000160959
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr9_-_7257877 | 0.03 |
ENSDART00000153514
|
si:ch211-121j5.4
|
si:ch211-121j5.4 |
| chr8_-_29706882 | 0.03 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr5_-_61431809 | 0.03 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr24_-_36271352 | 0.03 |
ENSDART00000153682
ENSDART00000155892 |
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr6_+_72040 | 0.03 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr4_-_5317006 | 0.03 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr4_-_67980261 | 0.03 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr18_+_33468099 | 0.03 |
ENSDART00000131769
|
v2rh9
|
vomeronasal 2 receptor, h9 |
| chr25_-_3087556 | 0.03 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr16_-_9383629 | 0.03 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr22_+_4947923 | 0.03 |
ENSDART00000135639
|
si:ch1073-104i17.1
|
si:ch1073-104i17.1 |
| chr3_+_25825043 | 0.03 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr6_+_42587637 | 0.03 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr15_-_37921998 | 0.03 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr17_-_28097760 | 0.02 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr9_+_51180508 | 0.02 |
ENSDART00000138990
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr8_-_44015210 | 0.02 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr3_+_26064091 | 0.02 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxc1a+foxc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.3 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 0.3 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.2 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.0 | 0.1 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.1 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.2 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.4 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 1.1 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.3 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 1.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.3 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |