Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
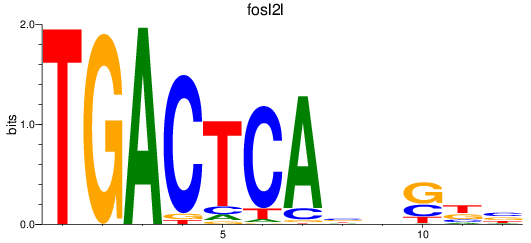
Results for fosl2l
Z-value: 1.28
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-59d15.4 | dr11_v1_chr20_+_9211237_9211237 | -0.49 | 1.8e-01 | Click! |
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_8671229 | 2.74 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr24_-_33366188 | 2.73 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr20_+_35445462 | 2.54 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr19_-_7043355 | 2.29 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr25_+_5972690 | 2.21 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr22_+_23359369 | 2.14 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr3_+_24708685 | 2.05 |
ENSDART00000153507
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr16_+_48714048 | 1.95 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr22_+_26600834 | 1.94 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr15_+_42235449 | 1.89 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr15_-_23442891 | 1.81 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr25_-_13408760 | 1.74 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr16_-_31598771 | 1.71 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr10_-_32877348 | 1.70 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr21_+_20901505 | 1.66 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr12_+_46582727 | 1.65 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr5_-_20678300 | 1.63 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr1_+_46598502 | 1.62 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr8_-_22288004 | 1.61 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr8_-_22288258 | 1.59 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr22_+_17828267 | 1.54 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr5_-_49951106 | 1.49 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr6_+_51713076 | 1.45 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr23_+_42254960 | 1.44 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr17_+_32623931 | 1.43 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr23_-_37113396 | 1.42 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr3_-_24205339 | 1.40 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr12_-_35787801 | 1.40 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr4_+_5317483 | 1.38 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr23_-_24226533 | 1.36 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr24_+_24726956 | 1.35 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr23_+_24989387 | 1.33 |
ENSDART00000172299
ENSDART00000145307 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr5_+_34549845 | 1.31 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr23_-_20010579 | 1.31 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr3_+_43086548 | 1.30 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr22_+_5574952 | 1.29 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr17_+_24597001 | 1.28 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr3_+_19687217 | 1.27 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr19_-_7070691 | 1.24 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr21_+_13245302 | 1.22 |
ENSDART00000189498
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr15_-_43978141 | 1.21 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr9_-_12493628 | 1.20 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr21_+_43199237 | 1.19 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr23_+_24272421 | 1.19 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr21_-_20945658 | 1.17 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr21_-_28640316 | 1.15 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr20_+_23501535 | 1.14 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr23_-_37113215 | 1.13 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr7_+_34549198 | 1.12 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr8_+_50190742 | 1.11 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr8_+_50534948 | 1.10 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr15_+_20352123 | 1.09 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr8_-_38201415 | 1.08 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr11_-_11965033 | 1.08 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr21_+_3901775 | 1.06 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr7_+_34549377 | 1.05 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr24_-_5691956 | 1.04 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr9_+_46745348 | 1.04 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr1_+_41690402 | 1.01 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr16_-_16226180 | 1.01 |
ENSDART00000108965
|
gra
|
granulito |
| chr14_+_23184517 | 0.98 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr19_-_5805923 | 0.94 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr19_+_15441022 | 0.94 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_27821 | 0.92 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr25_+_30196039 | 0.91 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr23_+_19655301 | 0.91 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr24_-_31090948 | 0.90 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr18_+_15841449 | 0.90 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr23_+_28128453 | 0.90 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr19_-_12078583 | 0.89 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr24_+_30215475 | 0.88 |
ENSDART00000164717
|
si:ch73-358j7.2
|
si:ch73-358j7.2 |
| chr1_+_46598764 | 0.88 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr25_+_16117138 | 0.88 |
ENSDART00000146604
|
far1
|
fatty acyl CoA reductase 1 |
| chr9_+_28140089 | 0.88 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr12_+_32292564 | 0.87 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr15_+_22867174 | 0.86 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr5_-_13076779 | 0.86 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr3_+_15776446 | 0.85 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr11_+_2596667 | 0.85 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr17_+_28624321 | 0.85 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr17_+_28675120 | 0.84 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr10_-_38443312 | 0.84 |
ENSDART00000141260
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr22_-_26265355 | 0.84 |
ENSDART00000169397
|
ccdc130
|
coiled-coil domain containing 130 |
| chr6_-_59357256 | 0.83 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr8_+_48965767 | 0.83 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr2_-_13691834 | 0.83 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr6_+_18367388 | 0.83 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr14_-_16807206 | 0.82 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr5_+_52706316 | 0.82 |
ENSDART00000169488
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr9_+_33154841 | 0.82 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr20_-_47347962 | 0.82 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr9_-_41025062 | 0.82 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr8_+_48966165 | 0.82 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr3_-_21062706 | 0.81 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr16_+_26547152 | 0.80 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr23_-_45398622 | 0.80 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr25_-_6432463 | 0.79 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr16_+_32136550 | 0.78 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr7_+_66884570 | 0.78 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr12_-_27212596 | 0.77 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr23_-_31693309 | 0.77 |
ENSDART00000146327
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr21_-_30254185 | 0.77 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr22_+_24603930 | 0.74 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr21_-_25612658 | 0.74 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr11_-_14102131 | 0.74 |
ENSDART00000085158
ENSDART00000191962 |
tmem259
|
transmembrane protein 259 |
| chr12_-_3077395 | 0.74 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_+_618005 | 0.73 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr9_+_33158191 | 0.72 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr14_+_29945070 | 0.72 |
ENSDART00000185039
|
fam149a
|
family with sequence similarity 149 member A |
| chr6_+_48348415 | 0.72 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr7_-_7764287 | 0.70 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr1_-_10577945 | 0.70 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr9_+_22388686 | 0.69 |
ENSDART00000182731
ENSDART00000181462 |
dgkg
|
diacylglycerol kinase, gamma |
| chr11_-_18601955 | 0.68 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr12_+_29173523 | 0.68 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr8_-_18516385 | 0.68 |
ENSDART00000149446
|
si:dkey-30h22.11
|
si:dkey-30h22.11 |
| chr7_-_26262978 | 0.68 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr15_-_23443588 | 0.68 |
ENSDART00000170427
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr21_+_17301790 | 0.67 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr21_+_3093419 | 0.67 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr21_-_14692119 | 0.67 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr20_-_47348116 | 0.66 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr9_+_2762270 | 0.66 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr4_-_20208166 | 0.66 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr9_+_22929675 | 0.66 |
ENSDART00000061299
|
tsn
|
translin |
| chr18_-_41160975 | 0.65 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr17_+_51743908 | 0.65 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr19_+_4139065 | 0.65 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr13_+_15800742 | 0.64 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr7_-_44704910 | 0.64 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr19_+_33093577 | 0.63 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr7_+_41147732 | 0.63 |
ENSDART00000185388
|
puf60b
|
poly-U binding splicing factor b |
| chr11_+_37905630 | 0.63 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr15_-_18197008 | 0.63 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr5_+_51227147 | 0.63 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr5_-_23464970 | 0.62 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr7_+_14005111 | 0.62 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr20_-_45040916 | 0.61 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr2_-_42827336 | 0.61 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr19_+_4051535 | 0.60 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr2_-_39675829 | 0.59 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr3_-_39190317 | 0.58 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr14_-_22100118 | 0.58 |
ENSDART00000157547
|
ssrp1a
|
structure specific recognition protein 1a |
| chr9_+_33222911 | 0.57 |
ENSDART00000135387
|
si:ch211-125e6.14
|
si:ch211-125e6.14 |
| chr5_+_61508418 | 0.57 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr3_+_20001608 | 0.57 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr3_+_34121156 | 0.56 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr7_-_26125092 | 0.56 |
ENSDART00000079364
|
snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr15_+_24704798 | 0.55 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr19_+_48359259 | 0.55 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr6_-_33685325 | 0.55 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr4_-_4535189 | 0.55 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr12_-_5455936 | 0.54 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr8_+_10835456 | 0.54 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr11_-_45152702 | 0.54 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr9_-_33608427 | 0.54 |
ENSDART00000100849
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr10_-_105100 | 0.54 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr23_+_19759128 | 0.52 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr7_-_71456117 | 0.52 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr6_+_33885828 | 0.52 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr13_+_6086730 | 0.52 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr6_+_49901465 | 0.52 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr3_+_19245804 | 0.52 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr10_+_16225117 | 0.52 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr1_+_52137528 | 0.51 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr25_+_18583877 | 0.51 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr7_-_6468611 | 0.50 |
ENSDART00000147978
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr15_+_40188076 | 0.50 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr25_-_11378623 | 0.50 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr12_+_1286642 | 0.50 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr17_+_43946269 | 0.50 |
ENSDART00000017376
|
ktn1
|
kinectin 1 |
| chr6_-_25371196 | 0.50 |
ENSDART00000187291
|
PKN2 (1 of many)
|
zgc:153916 |
| chr4_-_789645 | 0.48 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr22_-_11954861 | 0.48 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
| chr19_+_33093395 | 0.48 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr15_+_29709382 | 0.47 |
ENSDART00000099956
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr19_-_7272921 | 0.47 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr1_-_40123943 | 0.47 |
ENSDART00000146917
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr7_+_67381912 | 0.47 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr5_-_64900552 | 0.46 |
ENSDART00000073963
|
si:ch211-236k19.2
|
si:ch211-236k19.2 |
| chr20_-_34670236 | 0.46 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr2_+_25378457 | 0.45 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr21_+_25068215 | 0.44 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr4_+_77948970 | 0.44 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr25_+_32496723 | 0.44 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr13_+_12671513 | 0.44 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr8_-_1255321 | 0.43 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr11_-_13126969 | 0.43 |
ENSDART00000169052
ENSDART00000190120 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr23_-_14990865 | 0.43 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr9_-_9705512 | 0.43 |
ENSDART00000149021
|
gpr156
|
G protein-coupled receptor 156 |
| chr5_-_8910703 | 0.43 |
ENSDART00000097221
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr12_+_46582168 | 0.42 |
ENSDART00000189402
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr5_-_42123794 | 0.41 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr21_+_22892836 | 0.41 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr4_+_77933084 | 0.40 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr9_+_29323163 | 0.40 |
ENSDART00000184908
|
oxgr1b
|
oxoglutarate (alpha-ketoglutarate) receptor 1b |
| chr21_+_6290566 | 0.40 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr17_+_51744450 | 0.40 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr6_-_60079551 | 0.40 |
ENSDART00000154753
|
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr24_-_40009446 | 0.40 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.4 | 1.2 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.3 | 1.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 1.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.3 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.7 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 2.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 2.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.9 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 2.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.0 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 1.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 0.9 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 1.1 | GO:0098773 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.2 | 1.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.6 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.6 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.4 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.1 | 0.6 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 1.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.8 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 2.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.8 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.7 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.4 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 1.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.7 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.3 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.8 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.5 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 3.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 1.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 1.2 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.7 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.9 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.9 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 1.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.7 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.4 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.6 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 1.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 0.8 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.7 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.2 | 0.9 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.9 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 0.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 1.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.0 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 2.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 2.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 2.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 2.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 2.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.5 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 2.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |