Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
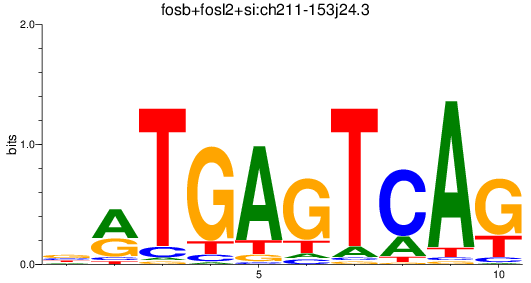
Results for fosb+fosl2+si:ch211-153j24.3
Z-value: 0.86
Transcription factors associated with fosb+fosl2+si:ch211-153j24.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2
|
ENSDARG00000040623 | fos-like antigen 2 |
|
fosb
|
ENSDARG00000055751 | FBJ murine osteosarcoma viral oncogene homolog B |
|
si_ch211-153j24.3
|
ENSDARG00000068428 | si_ch211-153j24.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-153j24.3 | dr11_v1_chr20_+_46560258_46560258 | 0.48 | 1.9e-01 | Click! |
| fosb | dr11_v1_chr18_+_36770166_36770166 | 0.21 | 5.9e-01 | Click! |
| fosl2 | dr11_v1_chr17_+_41302660_41302660 | 0.12 | 7.7e-01 | Click! |
Activity profile of fosb+fosl2+si:ch211-153j24.3 motif
Sorted Z-values of fosb+fosl2+si:ch211-153j24.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_24681292 | 1.64 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr9_-_11560110 | 1.50 |
ENSDART00000176941
|
cryba2b
|
crystallin, beta A2b |
| chr3_-_61185746 | 1.48 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr13_+_24280380 | 1.37 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr6_-_13783604 | 1.30 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr9_-_11560427 | 1.27 |
ENSDART00000127942
ENSDART00000061442 |
cryba2b
|
crystallin, beta A2b |
| chr22_-_26595027 | 1.05 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr2_+_48288461 | 0.96 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr16_+_23403602 | 0.93 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr7_-_8417315 | 0.85 |
ENSDART00000173046
|
jac1
|
jacalin 1 |
| chr20_-_26532167 | 0.85 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr6_+_29791164 | 0.84 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr16_-_21785261 | 0.84 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr11_-_18253111 | 0.82 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr25_+_13191391 | 0.81 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr7_+_8543317 | 0.81 |
ENSDART00000114998
|
jac8
|
jacalin 8 |
| chr7_-_8490886 | 0.81 |
ENSDART00000159012
|
jac6
|
jacalin 6 |
| chr21_-_44512893 | 0.78 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr23_+_35714574 | 0.78 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr7_-_8504355 | 0.75 |
ENSDART00000173067
|
loc564660
|
hypothetical protein LOC564660 |
| chr21_+_22845317 | 0.74 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr19_-_103289 | 0.73 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr5_-_25582721 | 0.72 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr3_+_24197934 | 0.71 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr19_+_7567763 | 0.71 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr18_-_5595546 | 0.70 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr21_-_30545121 | 0.70 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr18_+_46382484 | 0.70 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr16_+_42829735 | 0.69 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr11_+_13630107 | 0.68 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr12_-_212843 | 0.68 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr13_+_50368668 | 0.67 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr13_+_23988442 | 0.67 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr19_+_5072918 | 0.67 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr20_+_26880668 | 0.67 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr22_-_15593824 | 0.66 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr20_-_26531850 | 0.66 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr2_+_30379650 | 0.65 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr1_-_22861348 | 0.62 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr7_-_8602864 | 0.61 |
ENSDART00000173291
|
jac2
|
jacalin 2 |
| chr7_-_8438657 | 0.61 |
ENSDART00000173054
|
si:dkeyp-32g11.8
|
si:dkeyp-32g11.8 |
| chr2_+_26179096 | 0.60 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr7_+_25033924 | 0.58 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr1_-_40914752 | 0.57 |
ENSDART00000113087
|
hmx1
|
H6 family homeobox 1 |
| chr16_+_42830152 | 0.57 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr2_-_50298337 | 0.57 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr23_-_39636195 | 0.56 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr9_+_22080122 | 0.56 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr25_+_21832938 | 0.56 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr12_+_25945560 | 0.55 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr10_+_4987766 | 0.55 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr7_-_8577190 | 0.54 |
ENSDART00000173174
|
jac3
|
jacalin 3 |
| chr25_-_22187397 | 0.53 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr25_-_173165 | 0.53 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr25_+_13191615 | 0.51 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr5_-_66028714 | 0.50 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr11_+_26609110 | 0.49 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr6_-_24143923 | 0.48 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr11_+_13629528 | 0.48 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr16_+_20915319 | 0.48 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr14_-_11456724 | 0.46 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr25_-_18249751 | 0.46 |
ENSDART00000153950
|
si:dkey-106n21.1
|
si:dkey-106n21.1 |
| chr7_+_23875269 | 0.46 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr6_+_56147812 | 0.44 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_+_23960744 | 0.44 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr4_-_6809323 | 0.43 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr12_+_2399130 | 0.43 |
ENSDART00000075179
|
vstm4b
|
V-set and transmembrane domain containing 4b |
| chr9_-_710896 | 0.42 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr1_+_14253118 | 0.42 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr1_-_33380340 | 0.40 |
ENSDART00000181515
|
cd99
|
CD99 molecule |
| chr20_-_42972599 | 0.40 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr14_+_33882973 | 0.40 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr9_-_9992697 | 0.39 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr11_+_21050326 | 0.39 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr2_-_985417 | 0.39 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr2_+_42236118 | 0.38 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr7_-_8515550 | 0.38 |
ENSDART00000111896
|
jac9
|
jacalin 9 |
| chr4_+_77735212 | 0.38 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr25_+_21324588 | 0.36 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr10_-_27566481 | 0.36 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr7_+_20298333 | 0.36 |
ENSDART00000023089
ENSDART00000131019 |
acadvl
|
acyl-CoA dehydrogenase very long chain |
| chr19_-_7420867 | 0.36 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr18_-_15771551 | 0.35 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr10_+_31222656 | 0.35 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr1_-_59411901 | 0.35 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr20_+_19066596 | 0.34 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr23_-_29376859 | 0.34 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr9_-_34915351 | 0.33 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr1_-_52498146 | 0.33 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr8_+_31119548 | 0.33 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr4_+_16715267 | 0.32 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr17_-_28707898 | 0.32 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr19_+_22216778 | 0.32 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr16_+_9762261 | 0.32 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr7_+_53541173 | 0.32 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr23_+_9067131 | 0.32 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr5_-_9073433 | 0.31 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr22_+_16497670 | 0.31 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr24_-_36826504 | 0.31 |
ENSDART00000112694
|
fam171a2b
|
family with sequence similarity 171, member A2b |
| chr22_-_5006801 | 0.30 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr8_-_20291922 | 0.30 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr20_-_25626198 | 0.30 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr5_+_38276582 | 0.30 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr5_-_66028371 | 0.30 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr1_+_41588170 | 0.29 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr17_-_2945592 | 0.29 |
ENSDART00000174899
|
C14orf132
|
chromosome 14 open reading frame 132 |
| chr14_+_49382180 | 0.29 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr4_-_76488854 | 0.29 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr25_-_22191733 | 0.28 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr20_+_19066858 | 0.28 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr2_-_17827983 | 0.28 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr19_-_33212023 | 0.27 |
ENSDART00000189209
|
trib1
|
tribbles pseudokinase 1 |
| chr13_-_37122217 | 0.27 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_+_59073436 | 0.27 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr18_+_14477740 | 0.27 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr10_-_13239367 | 0.26 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr23_-_30045661 | 0.26 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr2_-_17828279 | 0.26 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr7_+_36041509 | 0.26 |
ENSDART00000162850
|
irx3a
|
iroquois homeobox 3a |
| chr7_+_60551133 | 0.25 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr25_+_8955530 | 0.25 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr11_+_77526 | 0.25 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr23_+_3731375 | 0.25 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr8_+_44358443 | 0.25 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr13_+_49175947 | 0.25 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr17_-_20711735 | 0.24 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr5_+_15819651 | 0.24 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr7_-_52963493 | 0.24 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr24_-_6078222 | 0.23 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr10_+_26747755 | 0.23 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr1_+_2190714 | 0.23 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr9_+_41024973 | 0.23 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr1_+_45323142 | 0.23 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr21_+_5636008 | 0.23 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr5_+_38200807 | 0.23 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr1_-_47771742 | 0.22 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr11_+_24251141 | 0.22 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr13_+_18533005 | 0.22 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr3_-_48259289 | 0.22 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr4_-_13902188 | 0.22 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr10_+_31222433 | 0.22 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr4_-_19742300 | 0.21 |
ENSDART00000066964
ENSDART00000100952 |
hgfa
|
hepatocyte growth factor a |
| chr19_-_32641725 | 0.21 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr8_+_20950401 | 0.21 |
ENSDART00000136484
ENSDART00000131329 ENSDART00000144451 ENSDART00000144929 |
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr13_-_15982707 | 0.21 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr19_-_31402429 | 0.21 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr17_-_26868169 | 0.21 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr9_-_27805801 | 0.21 |
ENSDART00000140608
ENSDART00000114542 |
si:rp71-45g20.10
|
si:rp71-45g20.10 |
| chr16_-_28878080 | 0.21 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr1_+_8601935 | 0.20 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr5_-_43959972 | 0.20 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr10_+_25947946 | 0.20 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr11_+_11267829 | 0.20 |
ENSDART00000026814
ENSDART00000173346 ENSDART00000151926 |
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr3_+_24986145 | 0.20 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr3_-_32859335 | 0.20 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr14_-_46117345 | 0.20 |
ENSDART00000172166
|
NDUFC1
|
si:ch211-235e9.6 |
| chr3_-_38918697 | 0.20 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr15_-_37733238 | 0.20 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr12_-_19250854 | 0.20 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr24_+_41940299 | 0.19 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr2_+_7849890 | 0.19 |
ENSDART00000114241
|
si:ch211-38m6.6
|
si:ch211-38m6.6 |
| chr25_-_3549321 | 0.18 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr22_+_25049563 | 0.18 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr15_-_31177324 | 0.18 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr19_+_21919856 | 0.18 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr9_+_38372216 | 0.18 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr21_-_30415524 | 0.18 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr9_-_21838045 | 0.18 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr18_+_5137241 | 0.18 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr23_-_437467 | 0.18 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr12_-_49154934 | 0.17 |
ENSDART00000153363
|
hmx3b
|
H6 family homeobox 3b |
| chr1_-_22735924 | 0.17 |
ENSDART00000134831
|
prom1b
|
prominin 1 b |
| chr1_-_52497834 | 0.17 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_+_49005321 | 0.17 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr10_+_1638876 | 0.17 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr9_+_7030016 | 0.17 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr8_+_28695914 | 0.16 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr1_+_31113951 | 0.16 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr11_+_11267493 | 0.16 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr8_+_43053519 | 0.16 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr4_-_77506362 | 0.16 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr25_+_13498188 | 0.15 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr8_+_4333914 | 0.15 |
ENSDART00000137528
|
myl2b
|
myosin, light chain 2b, regulatory, cardiac, slow |
| chr1_-_59139599 | 0.15 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr4_-_69189894 | 0.15 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr23_+_40460333 | 0.15 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr20_+_7084154 | 0.15 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr17_+_21452258 | 0.15 |
ENSDART00000157098
|
pla2g4f.1
|
phospholipase A2, group IVF, tandem duplicate 1 |
| chr18_+_31410652 | 0.15 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr12_+_33151246 | 0.15 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr5_+_9037650 | 0.15 |
ENSDART00000158226
|
si:ch211-155m12.1
|
si:ch211-155m12.1 |
| chr14_+_36869273 | 0.15 |
ENSDART00000111674
|
F2RL3
|
si:ch211-132p1.2 |
| chr5_+_40224938 | 0.14 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr1_-_40227166 | 0.14 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr17_+_15413412 | 0.14 |
ENSDART00000149871
|
cx40.8
|
connexin 40.8 |
| chr23_-_28294763 | 0.14 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr8_-_38105053 | 0.14 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr2_-_21847935 | 0.14 |
ENSDART00000003940
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr10_-_4316015 | 0.14 |
ENSDART00000109916
ENSDART00000180867 |
grin3a
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
| chr12_-_45669050 | 0.14 |
ENSDART00000124455
|
fbxl15
|
F-box and leucine-rich repeat protein 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosb+fosl2+si:ch211-153j24.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 0.7 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 0.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.4 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.7 | GO:0046638 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.6 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 1.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.5 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.6 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.7 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.0 | 0.7 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 1.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 4.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.4 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.6 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |