Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
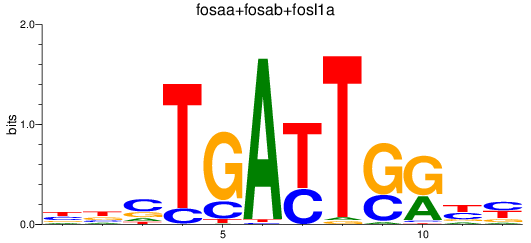
Results for fosaa+fosab+fosl1a
Z-value: 3.25
Transcription factors associated with fosaa+fosab+fosl1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl1a
|
ENSDARG00000015355 | FOS-like antigen 1a |
|
fosab
|
ENSDARG00000031683 | v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
|
fosaa
|
ENSDARG00000040135 | v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fosab | dr11_v1_chr20_-_46554440_46554440 | -0.78 | 1.3e-02 | Click! |
| fosaa | dr11_v1_chr17_-_50234004_50234004 | -0.77 | 1.6e-02 | Click! |
| fosl1a | dr11_v1_chr14_-_30747686_30747686 | -0.71 | 3.1e-02 | Click! |
Activity profile of fosaa+fosab+fosl1a motif
Sorted Z-values of fosaa+fosab+fosl1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_60886984 | 14.25 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_-_33647138 | 5.40 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr23_-_893547 | 5.26 |
ENSDART00000136805
|
rbm10
|
RNA binding motif protein 10 |
| chr19_+_14109348 | 5.22 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr11_+_29537756 | 5.00 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr9_+_29548195 | 4.74 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr13_-_4848889 | 4.24 |
ENSDART00000165259
|
mcu
|
mitochondrial calcium uniporter |
| chr24_+_10027902 | 4.19 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr24_-_10006158 | 3.91 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr9_+_29548630 | 3.89 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr16_+_54641230 | 3.77 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr20_-_52939501 | 3.66 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr24_+_12835935 | 3.57 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr2_-_17115256 | 3.32 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr3_+_7808459 | 3.22 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr24_-_9997948 | 3.03 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_9989634 | 3.01 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr7_+_30626378 | 3.00 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr12_-_22238004 | 3.00 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr13_+_8892784 | 2.99 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr1_+_30723380 | 2.98 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr21_+_3897680 | 2.97 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr1_+_30723677 | 2.96 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr12_+_48340133 | 2.96 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr14_-_35892767 | 2.93 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr18_+_14307059 | 2.88 |
ENSDART00000186558
|
zgc:173742
|
zgc:173742 |
| chr1_+_55476002 | 2.82 |
ENSDART00000152356
|
si:dkey-9c18.3
|
si:dkey-9c18.3 |
| chr24_-_9979342 | 2.80 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr7_-_73845736 | 2.74 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr22_-_10539180 | 2.70 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr1_-_18811517 | 2.64 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr25_+_36292465 | 2.62 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr19_-_2115040 | 2.62 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr25_-_17918536 | 2.58 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr8_-_2616326 | 2.56 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr3_-_60711127 | 2.53 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr5_-_54712159 | 2.53 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr12_-_42368296 | 2.53 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr8_-_20230559 | 2.52 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr20_-_2641233 | 2.51 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr16_+_48714048 | 2.45 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr17_+_25563979 | 2.45 |
ENSDART00000045615
ENSDART00000183162 |
qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr16_-_13680692 | 2.41 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr25_+_7435291 | 2.40 |
ENSDART00000172567
ENSDART00000163017 |
prc1a
|
protein regulator of cytokinesis 1a |
| chr14_-_30945515 | 2.37 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr21_-_32781612 | 2.35 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr10_-_39283883 | 2.32 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr3_+_46559639 | 2.26 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr10_+_6010570 | 2.22 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr14_-_22495604 | 2.22 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr24_-_9991153 | 2.22 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr16_-_24832038 | 2.22 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr3_-_40054615 | 2.20 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr25_-_17918810 | 2.19 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr22_-_5171362 | 2.17 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr7_-_48251234 | 2.16 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr10_-_25543227 | 2.15 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr18_-_25051846 | 2.15 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr10_-_32877348 | 2.14 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr25_-_6261693 | 2.14 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr13_+_2394264 | 2.13 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr16_+_50434668 | 2.09 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr7_-_6445129 | 2.08 |
ENSDART00000172825
|
FP325123.2
|
Histone H3.2 |
| chr8_+_8671229 | 2.03 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr20_+_27749133 | 2.03 |
ENSDART00000089013
|
vrtn
|
vertebrae development associated |
| chr7_+_34794829 | 2.03 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr21_-_43527198 | 2.01 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr14_+_21754521 | 1.97 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr9_-_40683722 | 1.97 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr10_+_39283985 | 1.96 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr19_+_791538 | 1.94 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr25_+_36292057 | 1.93 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr7_+_57088920 | 1.93 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr25_+_18436301 | 1.92 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr7_+_5905091 | 1.92 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr8_-_20230802 | 1.91 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr18_+_45573416 | 1.91 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr11_+_45287541 | 1.91 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr3_+_1167026 | 1.91 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr3_+_14571813 | 1.90 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr13_+_46941930 | 1.89 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_-_45029041 | 1.89 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr6_-_7686594 | 1.89 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr5_+_45139196 | 1.88 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr13_-_14929236 | 1.88 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr19_+_9111550 | 1.87 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr18_+_3243292 | 1.86 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr21_-_41624697 | 1.84 |
ENSDART00000100039
|
pcyox1l
|
prenylcysteine oxidase 1 like |
| chr3_-_26191960 | 1.82 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr12_+_13344896 | 1.82 |
ENSDART00000089017
|
rnasen
|
ribonuclease type III, nuclear |
| chr1_-_9527200 | 1.81 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr12_+_1469090 | 1.81 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr14_-_46374870 | 1.77 |
ENSDART00000185803
ENSDART00000188313 ENSDART00000031498 |
ccna2
|
cyclin A2 |
| chr19_-_35450857 | 1.77 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr12_+_1469327 | 1.77 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr9_+_45227028 | 1.76 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr2_-_17492080 | 1.74 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr16_+_45930962 | 1.73 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr5_-_3991655 | 1.73 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr7_+_17953589 | 1.73 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr12_+_46543572 | 1.73 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr7_+_51805525 | 1.72 |
ENSDART00000026571
|
slc38a7
|
solute carrier family 38, member 7 |
| chr22_-_5171829 | 1.72 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr2_-_17114852 | 1.71 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr3_-_6719232 | 1.71 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr1_-_54972170 | 1.70 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr1_-_54971968 | 1.69 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr7_-_33868903 | 1.69 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr22_-_16270462 | 1.69 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr17_-_26507289 | 1.69 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr13_+_11829072 | 1.67 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr17_+_23556764 | 1.67 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr3_+_18840810 | 1.66 |
ENSDART00000181137
ENSDART00000128626 ENSDART00000133332 |
tmem104
|
transmembrane protein 104 |
| chr1_+_54069450 | 1.65 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr5_+_56119975 | 1.65 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr5_+_30596822 | 1.64 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr5_+_30596477 | 1.63 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr24_+_17334682 | 1.63 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr15_+_15258758 | 1.63 |
ENSDART00000155719
|
c2cd3
|
C2 calcium-dependent domain containing 3 |
| chr25_-_6389713 | 1.61 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr19_+_32456974 | 1.61 |
ENSDART00000088265
|
atxn1a
|
ataxin 1a |
| chr20_+_13883131 | 1.61 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr19_-_15420678 | 1.60 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr5_+_30596632 | 1.58 |
ENSDART00000051414
|
hinfp
|
histone H4 transcription factor |
| chr6_-_1820606 | 1.58 |
ENSDART00000183228
|
FO834857.1
|
|
| chr7_+_25126629 | 1.57 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr3_-_70782 | 1.57 |
ENSDART00000110602
|
zgc:165518
|
zgc:165518 |
| chr22_+_10543329 | 1.57 |
ENSDART00000091850
|
atrip
|
ATR interacting protein |
| chr2_-_38117538 | 1.57 |
ENSDART00000013676
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr16_+_10329701 | 1.56 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr14_-_8903435 | 1.56 |
ENSDART00000160584
|
zgc:153681
|
zgc:153681 |
| chr7_-_51775688 | 1.56 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr4_-_1720648 | 1.56 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr24_+_21187027 | 1.56 |
ENSDART00000111754
ENSDART00000156108 |
usf3
si:ch73-93k15.5
|
upstream transcription factor family member 3 si:ch73-93k15.5 |
| chr3_-_60589292 | 1.55 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr3_-_10751491 | 1.55 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr21_+_13383413 | 1.54 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr16_+_40043673 | 1.54 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr3_-_21062706 | 1.54 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr2_+_105748 | 1.54 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr3_+_14463941 | 1.54 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr7_-_26497947 | 1.53 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr16_+_11297703 | 1.53 |
ENSDART00000125158
|
znf574
|
zinc finger protein 574 |
| chr22_-_4439311 | 1.52 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr13_-_51846224 | 1.52 |
ENSDART00000184663
|
LT631684.2
|
|
| chr15_+_42235449 | 1.51 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr10_+_5268054 | 1.51 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_-_6431158 | 1.51 |
ENSDART00000173199
|
si:ch1073-153i20.5
|
si:ch1073-153i20.5 |
| chr10_-_35257458 | 1.50 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr7_+_36467315 | 1.49 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr20_+_35438300 | 1.49 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr14_+_32942063 | 1.48 |
ENSDART00000187705
|
lnx2b
|
ligand of numb-protein X 2b |
| chr5_-_14500622 | 1.48 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr13_+_28618086 | 1.47 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr21_-_5077715 | 1.47 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr10_+_42542517 | 1.46 |
ENSDART00000005496
|
kctd9b
|
potassium channel tetramerization domain containing 9b |
| chr17_-_11418513 | 1.45 |
ENSDART00000064412
ENSDART00000151847 |
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr10_-_25699454 | 1.45 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr5_-_23574234 | 1.45 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr14_-_40850481 | 1.44 |
ENSDART00000173236
|
elf1
|
E74-like ETS transcription factor 1 |
| chr16_-_39241801 | 1.44 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr22_+_15960005 | 1.44 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr12_-_33659328 | 1.44 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr10_-_33379850 | 1.44 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_+_9326234 | 1.43 |
ENSDART00000104536
|
chsy1
|
chondroitin sulfate synthase 1 |
| chr3_+_16722014 | 1.42 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr22_+_32228882 | 1.42 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr14_+_31529958 | 1.42 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr16_-_46579936 | 1.42 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr7_-_33684632 | 1.42 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr2_-_40889465 | 1.41 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr14_-_14659023 | 1.41 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr12_+_5048044 | 1.41 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr3_-_11828206 | 1.41 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr5_+_45138934 | 1.41 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr20_+_13883353 | 1.40 |
ENSDART00000188006
|
nek2
|
NIMA-related kinase 2 |
| chr1_+_6135176 | 1.39 |
ENSDART00000092324
ENSDART00000179970 |
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr15_+_47418565 | 1.39 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr15_+_38299563 | 1.39 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr1_+_44523516 | 1.38 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr12_-_33359052 | 1.38 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr7_-_51300277 | 1.38 |
ENSDART00000174174
|
gc2
|
guanylyl cyclase 2 |
| chr21_-_41624489 | 1.38 |
ENSDART00000182365
|
pcyox1l
|
prenylcysteine oxidase 1 like |
| chr25_-_8625601 | 1.38 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr12_-_33582382 | 1.38 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr17_-_53329704 | 1.37 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr25_-_32363341 | 1.37 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr19_+_42432625 | 1.37 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr13_+_13945218 | 1.36 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr11_-_45152702 | 1.36 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr3_+_4997545 | 1.35 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr20_+_4157815 | 1.35 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr13_-_18691041 | 1.35 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr6_-_47796989 | 1.35 |
ENSDART00000136726
|
magi3b
|
membrane associated guanylate kinase, WW and PDZ domain containing 3b |
| chr9_+_38524881 | 1.35 |
ENSDART00000131846
|
osbpl11
|
oxysterol binding protein-like 11 |
| chr6_+_54687495 | 1.34 |
ENSDART00000189514
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr22_+_15959844 | 1.34 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr21_-_45363871 | 1.34 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr13_-_42536642 | 1.33 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr18_+_50961953 | 1.33 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosaa+fosab+fosl1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.9 | 2.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.9 | 4.6 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.9 | 2.6 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.8 | 2.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.8 | 6.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.8 | 3.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.7 | 3.0 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.7 | 2.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.7 | 3.7 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.7 | 2.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.7 | 5.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.7 | 4.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.7 | 2.7 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.6 | 3.2 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.6 | 1.9 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.6 | 2.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.6 | 0.6 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.5 | 3.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 4.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 2.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.5 | 6.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.5 | 0.9 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.4 | 2.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.4 | 5.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.4 | 1.7 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.4 | 2.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.4 | 1.2 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.4 | 2.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 2.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.4 | 1.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.4 | 1.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 1.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.4 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 2.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 0.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 1.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.4 | 3.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.3 | 0.7 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.3 | 2.4 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.3 | 1.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 0.9 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 2.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 0.9 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 10.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.3 | 3.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.3 | 0.9 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.3 | 0.9 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 3.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.3 | 2.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 1.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.3 | 5.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.3 | 1.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 0.8 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 0.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.3 | 1.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.3 | 3.0 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 1.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 | 1.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 0.7 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 1.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.2 | 0.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 0.9 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.2 | 0.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 1.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 0.7 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.6 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 1.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 6.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.2 | 0.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 3.7 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.2 | 0.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.6 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.7 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.2 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 1.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 1.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.8 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.2 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 2.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 7.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.2 | 1.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 0.6 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.2 | 1.1 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.2 | 1.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 1.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 1.1 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.2 | 0.4 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.1 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.5 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 0.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.7 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 2.6 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.2 | 2.0 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.0 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 0.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 1.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.5 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 0.8 | GO:0061162 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 0.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.9 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 1.1 | GO:0018027 | histone H3-K36 methylation(GO:0010452) peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.6 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 2.8 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.6 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 0.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.7 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 1.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 2.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.5 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 1.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.8 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.8 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.4 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.7 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 2.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 7.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 1.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 1.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 1.0 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 1.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 1.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.8 | GO:2001270 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.8 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.9 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 2.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 2.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.7 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.7 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.0 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 1.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 1.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 1.1 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 2.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.4 | GO:0043039 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.5 | GO:0014068 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 1.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.0 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.9 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.8 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 1.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.8 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 5.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 2.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.3 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.5 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.8 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) |
| 0.1 | 1.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.2 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 1.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.4 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.2 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.8 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 2.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.5 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 2.7 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 0.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 2.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.6 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.5 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.5 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.1 | 1.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 2.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 3.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte migration(GO:0002686) negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 1.3 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.8 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.5 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.1 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 3.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 6.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 2.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.7 | GO:0072531 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine-containing compound transmembrane transport(GO:0072531) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 3.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 2.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 3.7 | GO:0034249 | negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.2 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 3.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0098751 | osteoclast development(GO:0036035) bone cell development(GO:0098751) regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.7 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 3.1 | GO:0000819 | sister chromatid segregation(GO:0000819) |
| 0.0 | 1.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 6.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 2.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.6 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0043506 | regulation of JUN kinase activity(GO:0043506) |
| 0.0 | 0.1 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.9 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 1.2 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 2.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.5 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.2 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.8 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 1.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 1.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.6 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 1.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 2.8 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 3.3 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.2 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.5 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 1.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 4.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.2 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.4 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0002562 | somatic diversification of immune receptors(GO:0002200) somatic diversification of immune receptors via germline recombination within a single locus(GO:0002562) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 4.0 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 0.6 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0070640 | cellular response to nutrient(GO:0031670) vitamin D3 metabolic process(GO:0070640) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.9 | 2.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.7 | 3.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.7 | 2.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 4.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 1.8 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.5 | 3.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 3.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.8 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.4 | 1.7 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.4 | 1.7 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.4 | 2.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 1.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 2.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 1.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 3.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 3.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 3.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 4.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 0.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 18.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 8.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.2 | 0.5 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.2 | 1.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 2.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.8 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 1.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 2.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 3.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 2.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 5.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 18.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 0.5 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.6 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 2.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.7 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0017119 | cis-Golgi network(GO:0005801) Golgi transport complex(GO:0017119) |
| 0.0 | 1.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 7.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 1.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.7 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 6.7 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 1.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.0 | 4.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 1.0 | 5.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.8 | 2.3 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.7 | 2.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.7 | 2.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.7 | 2.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.6 | 3.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.6 | 1.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.6 | 5.5 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 1.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.5 | 1.8 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.4 | 1.8 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.4 | 2.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 1.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 1.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 1.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 1.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 1.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.4 | 1.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 2.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 2.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 2.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 2.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.3 | 0.9 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.3 | 0.9 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.0 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 0.9 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 2.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 1.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 2.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 1.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 0.8 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 1.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 0.6 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 1.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 2.8 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.8 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.5 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 0.7 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.5 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.2 | 0.5 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.2 | 1.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 2.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 2.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 5.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 2.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 2.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 2.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.4 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.4 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.7 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.4 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 1.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.4 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 3.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 4.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 17.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 18.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 1.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.1 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.8 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.6 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 3.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 2.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.8 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.0 | GO:0061608 | nucleocytoplasmic transporter activity(GO:0005487) nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 2.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 2.0 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 6.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 6.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.5 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 1.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 5.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 5.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 2.9 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 24.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 13.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 2.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 4.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 6.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 2.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.4 | 0.7 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.3 | 3.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 2.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 1.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 6.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 6.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 2.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 1.8 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.2 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 3.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 5.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 5.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 6.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 3.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |