Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
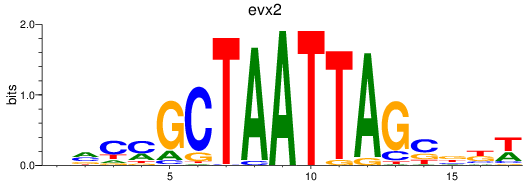
Results for evx2
Z-value: 1.03
Transcription factors associated with evx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
evx2
|
ENSDARG00000059255 | even-skipped homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| evx2 | dr11_v1_chr9_+_2002701_2002701 | -0.85 | 3.9e-03 | Click! |
Activity profile of evx2 motif
Sorted Z-values of evx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22313533 | 2.10 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr5_+_19933356 | 1.88 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr21_-_13661631 | 1.56 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr9_-_3934963 | 1.44 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr21_-_13662237 | 1.42 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr5_+_22067570 | 1.35 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr2_-_21819421 | 1.27 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr22_-_21897203 | 1.23 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr13_-_36663358 | 1.22 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr5_-_67629263 | 1.20 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_-_67241633 | 1.19 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr2_-_26720854 | 1.18 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr5_-_25733745 | 1.12 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr11_+_5588122 | 0.87 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr19_-_19379084 | 0.86 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr9_+_56194410 | 0.85 |
ENSDART00000168530
|
LO018176.1
|
|
| chr2_+_10007113 | 0.79 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr14_+_21820034 | 0.70 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr21_+_4410733 | 0.63 |
ENSDART00000136831
ENSDART00000131612 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr13_-_9091354 | 0.62 |
ENSDART00000140319
ENSDART00000142697 |
HTRA2 (1 of many)
|
si:dkey-112g5.13 |
| chr15_-_18115540 | 0.57 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr13_-_9111927 | 0.56 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr13_-_9236905 | 0.55 |
ENSDART00000142597
ENSDART00000147176 ENSDART00000181670 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-33c12.12 si:dkey-112g5.12 |
| chr18_+_7639401 | 0.54 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr10_+_34175476 | 0.53 |
ENSDART00000102123
|
zgc:174193
|
zgc:174193 |
| chr13_-_9045879 | 0.53 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr13_-_9257728 | 0.53 |
ENSDART00000145985
ENSDART00000169991 ENSDART00000132764 |
HTRA2 (1 of many)
|
zgc:173425 |
| chr13_-_9184132 | 0.51 |
ENSDART00000135289
ENSDART00000136309 ENSDART00000132630 |
HTRA2 (1 of many)
|
si:dkey-33c12.10 |
| chr13_+_9496748 | 0.51 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr13_+_9468535 | 0.50 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr21_+_4328348 | 0.45 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr13_+_9521629 | 0.42 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr17_-_5426458 | 0.41 |
ENSDART00000157937
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr13_-_9159852 | 0.39 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr13_-_9070754 | 0.37 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr9_+_1313418 | 0.37 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr21_+_4368735 | 0.35 |
ENSDART00000182817
|
CT027677.1
|
|
| chr21_+_4429001 | 0.32 |
ENSDART00000134520
|
HTRA2 (1 of many)
|
si:dkey-84o3.6 |
| chr13_-_9213207 | 0.31 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr21_+_4348600 | 0.27 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr24_-_35282568 | 0.22 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr2_-_1364678 | 0.19 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr19_+_47476908 | 0.15 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr4_+_11464255 | 0.03 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of evx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.6 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 1.3 | GO:0048796 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.9 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 1.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |